Complete Amino Acid Sequence of the Protease Inhibitor BWI-4a from Buckwheat Seeds
M. A. Belozersky1*, Y. E. Dunaevsky1, A. Kh. Musolyamov2, and T. A. Egorov2
1Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, Moscow, 119899 Russia; fax: (095) 939-3181; E-mail: mbeloz@genebee.msu.su2Shemyakin and Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, ul. Miklukho-Maklaya 16/10, Moscow, 117871 Russia; fax: (095) 330-7301; E-mail: ego@ibch.siobc.ras.ru
* To whom correspondence should be addressed.
Received April 26, 2000; Revision received July 1, 2000
The complete amino acid sequence of the protease inhibitor BWI-4a from buckwheat (Fagopyrum esculentum Moench) seeds has been established by automated Edman degradation in combination with MALDI-TOF mass spectrometry. The inhibitor molecule consists of 67 amino acid residues with a single disulfide bond. Its N-terminus is blocked by a pyroglutamic acid residue. The reactive site of the inhibitor contains an Arg43-Asp44 bond. Mass spectrometry revealed that inhibitor BWI-4a is present in buckwheat seeds in two isoforms differing by a single amino acid substitution of Gly40 for Ala40. Analysis of the amino acid sequence of the BWI-4a inhibitor indicates that this inhibitor is a member of the potato proteinase inhibitor I family.
KEY WORDS: amino acid sequence, buckwheat seeds, mass spectrometry,microheterogeneity, protease inhibitors
Abbreviations: MALDI-TOF) matrix assisted laser desorption ionization time-of-flight; NT and CT) N- and C-terminal fragments of BWI-4a, respectively; NS and CS) peptides obtained by digestion with V8 endoproteinase of NT- and CT-fragments of BWI-4a, respectively; RP) reversed-phase; TFA) trifluoroacetic acid.
Plant protease inhibitors continue to attract the attention of
researchers because of their increasing use in medicine and
biotechnology. Plant protease inhibitors have been studied much less
than similar proteins of animals, and their physiological role is not
understood conclusively. We have previously obtained and described two
groups of protease inhibitors from buckwheat seeds--anionic and
cationic--based on their behavior on ion exchangers [1]. While the anionic inhibitors suppressed growth and
development of pathogenic fungi and inhibited proteinases secreted by
these fungi, the cationic inhibitors effectively inhibited some
bacterial proteinases as well [1]. The amino acid
sequence of the anionic inhibitor BWI-1a was established [2]. Its sequence is similar to the sequence of one of
the two trypsin inhibitors described later by Pandya et al. [3]. The amino acid sequence of another trypsin
inhibitor from buckwheat seeds was elucidated by Park et al. [4]. In the present paper, we report the amino acid
sequence of the anionic protease inhibitor BWI-4a from the seeds of
buckwheat.
MATERIALS AND METHODS
Plant material. Dry seeds of buckwheat (Fagopyrum esculentum Moench) cv. Shatilovskaya-5 were used.
Isolation of the inhibitor. The inhibitor was isolated by a three-step procedure. First, the seed extract was subjected to affinity chromatography on a trypsin-Sepharose column as described previously [5]. The fraction containing the total preparation of buckwheat seed trypsin inhibitors was further chromatographed on a Mono Q HR 5/5 (Pharmacia, Sweden) column using an FPLC chromatograph (Pharmacia). The sample of the inhibitors (10 mg) was applied to the column in 20 mM K,Na-phosphate buffer, pH 6.8. The column was then washed with the same buffer, and the protein was eluted with a linear NaCl gradient (0-100 mM, 1 ml/min, 25 min). The preparation of the BWI-4a inhibitor was finally purified by reversed-phase (RP) chromatography on an Aquapore RP-300 (4.6 × 100 mm) column (Applied Biosystems, Inc., USA) with an acetonitrile gradient at a flow rate of 0.5 ml/min. Solvent A: 0.1% (v/v) TFA; solvent B: acetonitrile containing 0.08% (v/v) TFA.
Assay of the inhibitor activity. Five microliters of a trypsin solution (1 mg/ml) was added to the preparation of the inhibitor in 100 mM K,Na-phosphate (pH 6.8, 1-10 µl) and the mixture was incubated at room temperature for 10 min. The residual trypsin activity was assayed according to Erlanger et al. [6] with N-benzoyl-D,L-arginine-p-nitroanilide (0.2 mM) in 500 mM K,Na-phosphate (pH 7.5) as a substrate. One unit of the inhibitor activity produced a decrease in absorption of the solution by 0.1 unit in the trypsin activity assay.
Reduction and alkylation of the protein. The inhibitor preparation (0.5 mg) was reduced and alkylated with 4-vinylpyridine [7]. The alkylated protein was desalted by RP-HPLC and evaporated in a Speedvac apparatus (Savant, USA).
Digestion of the protein by V8 protease. Reduced and alkylated protein (300 µg) was dissolved in 100 µl of 100 mM NH4HCO3 (pH 7.8) and digested by 2 µg of Staphylococcus aureus V8 protease (Sigma, USA) for 16 h at 37°C. The resulting peptides were separated by RP-HPLC and the solvent evaporated using the Speedvac.
Amino acid analysis. The amino acid analysis of the inhibitor was carried out according to a standard procedure with a Hitachi 835 (Japan) amino acid analyzer. Cysteine was determined as cysteic acid after oxidation of the protein with a mixture of H2O2 and 88% performic acid (1 : 9 v/v), followed by hydrolysis with 5.7 N HCl (110°C, 22 h). Tryptophan was determined after hydrolysis of the protein with 4 M methanesulfonic acid containing 0.2% tryptamine.
Automated Edman degradation. Amino acid sequences of the protein and peptides were determined using a Model 816 Protein/Peptide Sequencer (Knauer, Germany) equipped with a Model 120A PTH-analyzer (Applied Biosystems). Preparations of the protein and peptides were dissolved in 10-15 µl of 50% acetonitrile containing 0.1% trichloroacetic acid and applied in 5 µl portions to an Immobilone P membrane (Millipore, USA) for sequencing.
Mass spectrometry. Mass spectra of the inhibitor and its fragments as well as the sequences of some peptides were determined with a MALDI-TOF Reflex II (Brucker-Franzen Analytik GmbH, Germany) mass spectrometer according to the protocols of the manufacturer.
RESULTS AND DISCUSSION
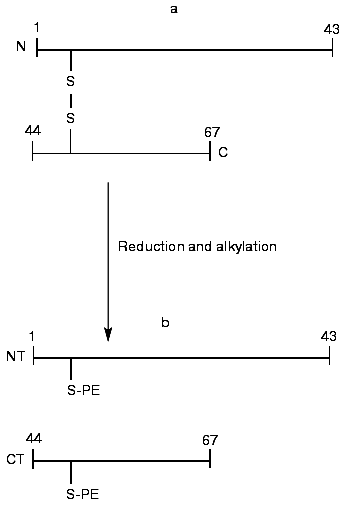
The BWI-4a protease inhibitor preparation was obtained by a previously developed procedure that included affinity chromatography on trypsin-Sepharose, ion-exchange chromatography, and RP-HPLC [5]. For determination of the sequence of the inhibitor, we took advantage of the fact that during affinity chromatography on trypsin-Sepharose a single peptide bond was split at its reactive site. Such cleavage often occurs by limited proteolysis of protease inhibitors with trypsin at acid pH [8]. Mass-spectrometric analysis of unreduced BWI-4a inhibitor showed the presence of two polypeptides with masses of 7463.3 and 7482.8 daltons (table). At the same time, the N-terminal sequence analysis of the inhibitor preparation by automated Edman degradation revealed only a single sequence corresponding to the N-terminal part of the C-terminal fragment (CT) and identical to the C-terminal part of the BWI-1a inhibitor (Fig. 1). These data indicated, first, that BWI-4a has a blocked N-terminal amino acid residue and, second, that one peptide bond at the reactive site of the inhibitor was split, presumably at the carboxyl side of the arginine residue, during affinity chromatography on trypsin-Sepharose, as demonstrated earlier for the buckwheat seed BWI-1a inhibitor [2]. Thus, the inhibitor preparation consisted of two polypeptide chains connected by a single disulfide bond (Fig. 1, table). Reduction and alkylation of BWI-4a with 4-vinylpyridine resulted in formation of two fragments--N-terminal (NT) and C-terminal (CT) (Fig. 1, table)--which were separated by RP-HPLC and subjected to Edman sequencing. The results of sequencing of these fragments are presented in the table.
Summary of the structural analysis of BWI-4a inhibitor from buckwheat
seeds. <E, pyroglutamic acid residue. Cysteine residues in the
unreduced BWI-4a form disulfide bonds and those of the NT- and
CT-fragments are alkylated with 4-vinylpyridine. Cysteine residues and
amino acid residues differing between two inhibitor components are
shown in bold. Amino acid sequences of peptides were determined by
Edman degradation and mass spectrometry. The amino acid sequences of
peptides determined only by Edman degradation are underlined, and those
identified only by mass spectrometry are shown in italic
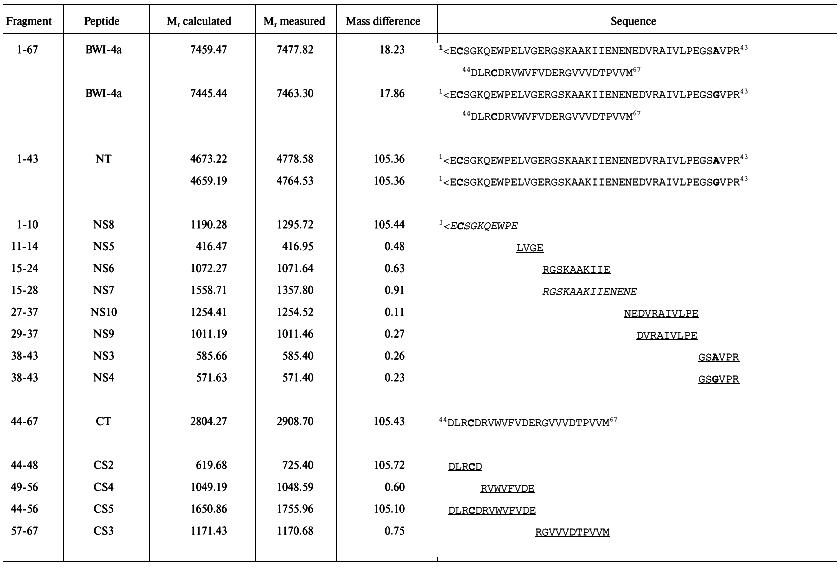
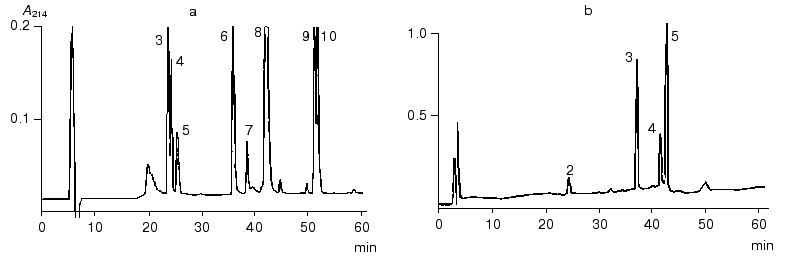
The N-terminal sequence of both the NT-fragment and the intact inhibitor could not be determined by direct Edman degradation due to the blockage of its N-terminal amino acid residue. The mass spectra of the NT-fragment indicated the presence of two components with masses of 4764.5 and 4778.6 daltons, differing in mass by 14.1 daltons. To determine the structure of the NT-fragment, it was subjected to digestion by V8 protease. As a result, 8 peptides (NS3, NS4, NS5, NS6, NS7, NS8, NS9, and NS10) were obtained (table), separated by RP-HPLC (Fig. 2), and analyzed. The sequences of peptides NS3, NS4, NS5, NS6, NS9, and NS10 were determined by Edman degradation in combination with mass spectrometry, and the structure of peptides NS7 and NS8 by mass spectrometry only. Mass-spectrometric analysis of the structure of peptide NS8 revealed that its N-terminal residue was the pyroglutamic acid residue. It is noteworthy that the presence of a pyroglutamic amino acid residue at the N-terminus of protease inhibitors was reported previously for carboxypeptidase inhibitors from potato [9] and tomato [10]. It was established that two peptides, NS3 and NS4, differed by only one amino acid residue (Ala for Gly, respectively). This was in good agreement with the results for mass analysis of peptides NS3 and NS4, which differed in mass by 14.00 daltons, corresponding to the mass difference between Ala and Gly.Fig. 1. Structure of the BWI-4a protease inhibitor from buckwheat seeds before (a) and after (b) reduction and alkylation. PE, ethylpyridine.
To determine the sequence of the CT-fragment (24 residues) of the BWI-4a inhibitor, it was subjected to automated Edman degradation, and the amino acid sequence from Asp44 to Val59 was established. To determine the complete structure of the CT-fragment, it was hydrolyzed with V8 protease, and the resulting peptides CS2, CS4, CS3, and CS5 were separated by RP-HPLC (Fig. 2). Amino acid sequences of these peptides were established by direct Edman degradation in combination with mass spectrometry. Analysis of the sequences of all of the peptides resulted in the determination of the complete sequence of the CT-fragment of BWI-4a inhibitor (table).Fig. 2. RP-HPLC elution profiles of peptides obtained after hydrolysis of BWI-4a inhibitor by V8 protease: a) peptides obtained after hydrolysis of the N-terminal fragment; b) peptides obtained after hydrolysis of the C-terminal fragment.
Thus, determination of the amino acid sequence of the NT- and CT-fragments of BWI-4a protease inhibitor, carried out by automated Edman degradation and mass spectrometry, revealed that the molecule of the inhibitor consists of 67 amino acid residues. The N-terminus of the molecule is blocked by a pyroglutamic acid residue. The BWI-4a inhibitor is present in buckwheat seeds in two isoforms, BWI-4a´ and BWI-4a´´, differing in the nature of the amino acid residue at position 40 (table).
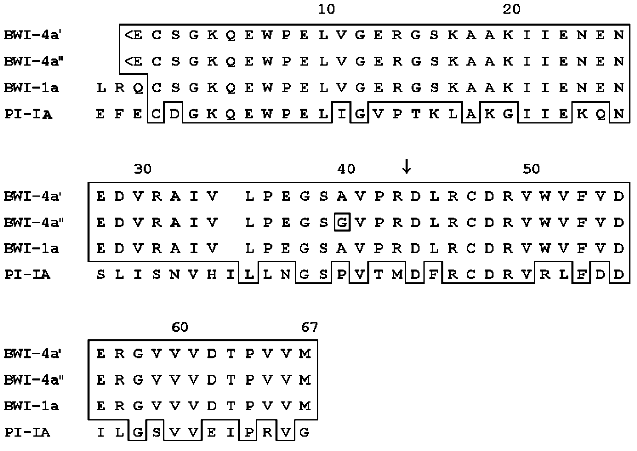
Computer analysis of the amino acid sequence of the BWI-4a buckwheat seed protease inhibitor revealed its homology with potato proteinase inhibitor PI-IA and buckwheat seed inhibitor BWI-1a (Fig. 3). These data indicate that BWI-4a is also a member of the potato proteinase inhibitor I family, based on the structure of proteinase inhibitors PI-I from potato [11]. According to homology of proteinase inhibitors of this family and BWI-4a inhibitor, it can be concluded that the reactive site of BWI-4a contains the Arg43-Asp44 amino acid residues (Fig. 3). The inhibitor molecule is stabilized by a single disulfide bond formed between Cys2 and Cys47.
It has been demonstrated previously that buckwheat seed BWI-4a protease inhibitor is able to suppress the activity of trypsin only, whereas BWI-1a inhibitor inhibits both trypsin and chymotrypsin [5]. Based on the data obtained in the present work, it can be proposed that the inability of BWI-4a inhibitor to inhibit chymotrypsin results from the difference in the N-terminal parts of these inhibitors, expressed in the absence of Leu and Arg and the presence of a pyroglutamic acid residue at the N-terminus of BWI-4a.Fig. 3. Comparison of amino acid sequences of buckwheat seed protease inhibitors BWI-4a and BWI-1a and potato proteinase inhibitor PI-IA [12]. Identical amino acids in the sequences are boxed. The arrow indicates the amino acid residue at the P1 position at the active site. <E, pyroglutamyl.
The authors are very grateful to Dr. Uwe Rapp (Brucker-Franzen Analytik GmbH, Bremen, Germany) for help with mass spectrometry of proteins and peptides.
This work was supported by grants from the Russian Foundation for Basic Research and State Programs “Phytobiotechnology” and “Basic Life Sciences”.
REFERENCES
1.Dunaevsky, Y. E., Pavlukova, E. B., Beliakova, G.
A., Tsybina, T. A., Gruban, T. N., and Belozersky, M. A. (1998) J.
Plant Pysiol., 152, 696-702.
2.Belozersky, M. A., Dunaevsky, Y. E., Musolyamov, A.
X., and Egorov, T. A. (1995) FEBS Lett., 37, 264-266.
3.Pandya, M. J., Smith, D. A., Yarwood, A., Gilroy,
J., and Richardson, M. (1996) Phytochemistry,43,
327-331.
4.Park, S., Abe, K., Kimura, M., Urisu, A., and
Yamasaki, N. (1997) FEBS Lett., 400, 103-107.
5.Dunaevsky, Y. E., Pavlukova, E. B., and Belozersky,
M. A. (1996) Biochem. Mol. Biol. Int., 40, 199-208.
6.Erlanger, B. F., Kokowsky, N., and Cohen, W. (1961)
Arch. Biochem. Biophys., 95, 271-278.
7.Thomsen, J., and Bayne, S. J. (1988) J. Prot.
Chem., 7, 295-296.
8.Ozawa, K., and Laskowski, M. J. (1966) J. Biol.
Chem., 241, 3955-3960.
9.Hass, G. M., Nau, H., Bilmann, K., Grah, D. T.,
Ericsson, L., and Neurath, H. (1975) Biochemistry, 14,
1334-1341.
10.Hass, G. M., and Hermodson, M. A. (1981)
Biochemistry, 20, 2256-2261.
11.Mosolov, V. V., and Valueva, T. A. (1993)
Plant Protein Inhibitors of Proteolytic Enzymes [in Russian],
VINITI, Moscow.
12.Richardson, M., and Cossins, L. (1974) FEBS
Lett., 45, 11-13.