Dependence of Inorganic Polyphosphate Chain Length on the Orthophosphate Content in the Culture Medium of the Yeast Saccharomyces cerevisiae
V. M. Vagabov*, L. V. Trilisenko, and I. S. Kulaev
Skryabin Institute of Biochemistry and Physiology of Microorganisms, Russian Academy of Sciences, Pushchino, Moscow Region, 142292 Russia; fax: (7-095) 923-3602; E-mail: kulaev@ibpm.serpukhov.su* To whom correspondence should be addressed.
Received November 10, 1999
The content of inorganic linear polyphosphate (polyP) and the polymeric degree (n) of these compounds were determined in the process of growth of the yeast Saccharomyces cerevisiae VKM Y-1173 in a medium, which contained varying Pi amount with the constant level of all the necessary components. For this purpose, a combination of chemical methods of polyP extraction and 31P-NMR spectroscopy studies of their chain length were used. After 7 h of phosphate starvation, the yeast was shown to use almost completely the phosphate reserve in the form of polyP localized in various cell compartments to support their vitality. The polyP drop was followed by a considerable shortening of the polymer chain length of acid-soluble (polyP1) and two alkali-soluble (polyP3 and polyP4) fractions. Under the same conditions, the content of a salt-soluble fraction (polyP2) decreased almost 20-fold followed by a simultaneous increase of the chain length nearly 2-fold. As a result, fraction chain length ranged up to n = 40-45. Replacement of the yeast cells after phosphate starvation to a complete phosphate- and glucose-containing medium resulted in super-accumulation (“overcompensation”) of polyP within 2 h mainly in polyP3 and, to a lesser degree, in polyP1, polyP2, and polyP5 fractions. In polyP4 fraction localized as polyP3 at the cell surface, the polyP super-accumulation was not detected. The increase of polyP amount in the fractions mentioned turned out not to be accompanied by simultaneous elongation of their chain length and occurred at the lowest level that is characteristic of a polymer level for each fraction. Further cultivation of the yeast on the complete medium during 2 h had little or no effect on polyP content in the cells but led to elongation of polyP chain length especially in the polyP3 and polyP4 fractions. A phenomenon of considerable elongation of polyP chain length against the background of their fixed content revealed in the yeast growing on the complete medium suggests that these organisms possess a previously unknown discrete way of polyP biosynthesis, which results first in the formation of comparatively low-molecular-mass chains followed by that of high-molecular-mass polymers.
KEY WORDS: yeast, polyphosphates, overcompensation, 31P-NMR spectroscopy
Recent intensive studies of inorganic polyphosphates (polyP) have significantly extended our view on the functions of these compounds in the metabolism of microorganisms. PolyP were shown not only to act as a reserve reutilized under phosphate and energy deficiency, but may also be involved in the transport of particular ions and substances into cells, such as metal ions and sugars, in osmoregulation, in the biosynthesis of nucleic acids, and in the maintenance of the fine structural organization of membranes and cell wall [1-5].
It has been established recently that the metabolism of linear polyP localized in various cell compartments is closely associated with the basic processes occurring in these structures. For example, it has been demonstrated that synthesis of one of the polyP fractions localized in the cell wall is associated with that of mannoproteins, the main components of these structures [2, 6]. Investigation of enzymatic properties of polyP metabolism revealed their specificity to the chain length of the substrates attacked [7-9]. However, still now little is known about the structure of polyP localized in various cell compartments. Successive treatment of biomass with cold diluted perchloric acid, salt, and then weak alkali allowed the isolation from yeast cells of polyP with distinct chain length ranging from short (n = 2-8) to high polymers, the chain of the latter containing no less than two hundred orthophosphate residues (n = 200) [10, 11].
Our recent studies have shown that the chain length of different polyP fractions depends not only on the method of their fractionation but on the growth stage of the yeast culture under study as well. In the early logarithmic phase of growth on complete medium rich in carbon and phosphate, the degree of polyP polymerization falls drastically against a background of their active synthesis. This was observed for most polymeric fractions. The increase of the high-polymer polyP length was detected again only at the late stationary growth phase when the synthesis was stopped [12].
The aim of this work was to study the content and the degree of polymerization of polyP during the growth of yeast cells under conditions of Pi extremes in the cultivation medium.
MATERIALS AND METHODS
The yeast Saccharomyces cerevisiae VKM Y-1173 was grown in shaken flasks containing 200 ml of Reader medium [13] with some modifications. The medium (1 liter) was supplemented with 3 g (NH4)2SO4, 0.7 g MgSO4, 0.4 g Ca(NO3)2, 0.5 g NaCl, 1 g KH2PO4, 0.1 g K2HPO4, 0.25 mg (NH4)2SO4·FeSO4·6H2O, 20 g glucose, 2 g yeast extract, and 5 ml of microelement solution having the following composition: 50 mg KI, 28 mg H3BO3, 25 mg MnSO4·7H2O, 22 mg ZnCl2·7H2O, 19.5 mg CuSO4·5H2O, 12.5 mg Na2MoO4·2H2O. In phosphate-free medium, the orthophosphate salts were substituted for equimolar quantity of KCl, and instead of the yeast extract, which also contains phosphate, 2 mg/liter inositol was introduced. The yeast was grown at 29°C with shaking. The culture growth was examined with a photoelectrocolorimeter at 530 nm and cell thickness 3.07 mm. The cells were harvested by centrifugation at 3000g, washed twice with distilled water, transferred into flasks for polyP fractionation, and harvested under the conditions mentioned.
The isolation of separate polyP fractions followed the method of Langen and Liss in Kulaev et al. modification [14] by a sequential treatment of the yeast with acid, salt, and alkali solutions. As a result, the following polyP fractions were obtained: acid-soluble (polyP1), salt-soluble (polyP2), two alkali-soluble (polyP3 and polyP4), and polyP5 fractions detected by the appearance of orthophosphate after the hydrolysis of the biomass remainder in 0.5 M HClO4 at 90°C for 20 min after removal of the preceding fractions. To obtain more concentrated solution of polyP and, thus, to facilitate their precipitation with Ba2+ salts, the extractant to biomass ratio was varied from 5 to 0.5 ml/g wet weight depending on the polyP content in the fraction.
To study polyP by 31P-NMR spectroscopy, the polymers were precipitated from extracts obtained with a saturated solution of Ba(NO3)2, pH 8.2. Two polyP precipitates were obtained from the acid-soluble fraction, first at pH 4.5 and then at pH 8.2. The polyP precipitates were transformed to a soluble state by treating them with Dowex AG 50W×8 ion-exchange resin in NH4+ form. Surplus Ba2+ and other metals interfering with NMR measurements were chelated by adding 20 mM EDTA. The pH value of the solutions was adjusted to 7.2. PolyP concentration in the preparations varied from 150 to 1000 µg Pi/ml.
31P-NMR spectra were recorded on an AM-400 Fourier transform NMR spectrometer (Bruker, Germany) with a superconducting magnet operated at 116.98 MHz, 45° pulse, and 1 sec delay. The scan number was more than 1000. A solution of disodium salt of ethylenediaminephosphonic acid with a chemical shift of 12.8 ppm with respect to 85% H3PO4 was used as the standard.
The length of polyP fractions was calculated from the formula based on the ratio of the areas of 31P-NMR-spectral peaks corresponding to the internal and terminal phosphate groups [15].
Phosphate concentration was estimated according to Berenblum and Chain in the modification of Weil-Malherbe and Green [16]. Glucose was measured by the phenol-sulfuric acid method [17].
To determine dry weight, aliquots of cell suspensions were applied on filters preliminary brought to a constant weight by drying under vacuum at 85°C and dried to a constant weight under the same conditions.
The yeast extract and Dowex AG 50W×8 were purchased in Serva (Germany); other substances were of chemically pure grade (Reakhim, Russia).
RESULTS AND DISCUSSION
To elucidate the effect of Pi content in the medium on the accumulation and qualitative changes of polyP in S. cerevisiae cells, the scheme of their cultivation presented in the Fig. 1b is elaborated.
The yeast is first cultivated for 4 h in a complete Rider medium containing 9 mM Pi and 111 mM glucose as a carbon source (point A). Then the cells are placed on the same medium except depleted of Pi under sterile conditions and the culture is grown during 7 h (point B). Further, the cells are again placed on fresh complete medium, where the culture is growing for 2 and 4 h (points C and D, respectively). At the growth points mentioned, biomass aliquots are sampled under sterile conditions to analyze the content of different polyP fractions and to determine the polymer chain length with 31P-NMR spectroscopy as indicated above (see “Materials and Methods”). The polymer degree of polyP5 is not determined since its extraction conditions lead to polyP hydrolysis.
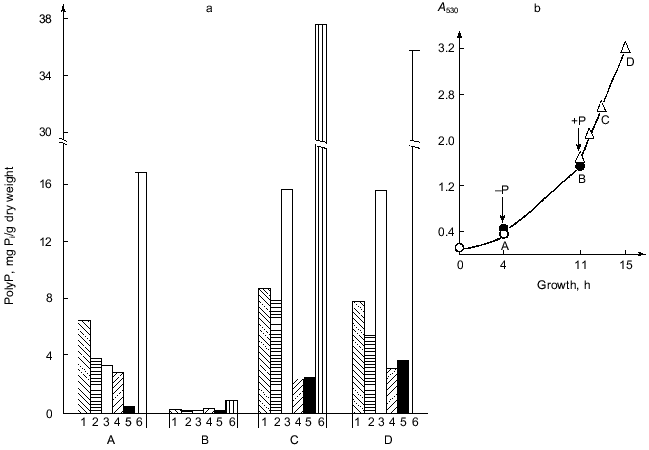
As seen from the growth curve at Fig. 1b, the first point A corresponds to the beginning of the logarithmic stage of cell growth. In a phosphate-free medium, the yeast biomass increases more than 3-fold during 7 h of growth (point B).Fig. 1. a) PolyP content in the cells of S. cerevisiae under different conditions of their cultivation: 1) polyP1; 2) polyP2; 3) polyP3; 4) polyP4; 5) polyP5; 6) polyP sum. b) Growth curve of the yeast as a function of Pi content in the medium. Point A) complete medium, transfer to a phosphate-free one; point B) phosphate-free medium, transfer to the complete one; points C and D) 2 and 4 h of growth on the complete medium.
It is known that various polyP fractions are localized in separate cell compartments. The low-polymer acid-soluble polyP1 fraction appears to be localized in volutin granules and vacuoles. The salt-soluble polyP2 fraction, the synthesis of which is coupled to that of nucleic acid, is possibly located in the nucleus; the most high-molecular-mass fractions, polyP3 and polyP4, are localized at the cell surface [1]. Nothing is yet known concerning the localization of the polyP5 fraction in the cells.
PolyP in various cell compartments act not only as phosphate storage but also have some other functions. In particular, the polyP3 fraction is involved in the maintenance of the fine structural organization of the yeast cell wall [4]. As is evident from the data presented, almost complete polyP utilization of absolutely all fractions is seen within 7 h of phosphate starvation. Placing of the yeast cells in fresh complete medium facilitates active polyP synthesis. As this takes place, the intensity of this polymer accumulation is not however the same for different fractions. After 2 h of growth (point C), the most active polyP synthesis occurs in the alkali-soluble polyP3 fraction, the content of which increased almost 400-fold. This suggests that the cells primarily restore normal function of the cell surface disrupted during the phosphate starvation. The increase in the activity of the enzymes of polyP synthesis in cells after their transfer from Pi-critical to complete medium (point C) leads to super-accumulation of polyP known as the phenomenon of “overcompensation” [10].
A comparison of polyP content in the yeast before phosphate starvation (point A) and after the transfer of the culture to the fresh complete medium (point C) indicates that super-accumulation is not characteristic for all polyP fractions. As seen from Fig. 1a, the maximal accumulation under the conditions in question is observed for the polyP3 (almost 5-fold) and polyP5 (more than 8-fold) fractions. The salt-soluble polyP2 fraction increases 2-fold, whereas the acid-soluble one increases less than 1.3-fold. At the same time, polyP4 does not even reach the initial level of polymers of this fraction at point A. These results suggest that the mechanism of synthesis of distinct polyP fractions may differ from one another [1, 6] or, at least, they are variously regulated in overcompensation. Further intensive culture growth in the Pi- and glucose-containing medium (point D) causes practically no change the polyP content. A small drop in the total polyP content of cells under these conditions is associated with the decrease of the polyP1 and polyP2 fractions. In this case the polyP4 and polyP5 fractions increase moderately (1.4- and 1.5-fold, respectively), and the content of the polyP3 fraction is practically unchanged. These data point to a peculiar kind of saturation or a balance of systems of the synthesis and degradation in all polyphosphate fractions of the yeast provided that the concentration of Pi and glucose in the medium is sufficient.
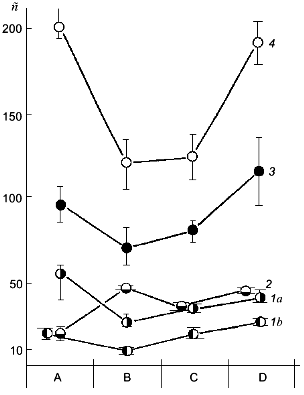
Against the background of our results on the change of polyP amount under simulated growth conditions, the changes in polyP chain length at different stages of this experiment are of a special interest (Fig. 2). This phenomenon has not been described previously. The degree of polymerization (n) of polyP fractions was estimated from the areas of 31P-NMR-spectral peaks. As seen from Fig. 2, the polyphosphates of the acid-soluble fraction can be separated into two sub-fractions by precipitation with barium salt first at pH 4.5 and then at pH 8.2. One of these sub-fractions (pH 8.2) contains rather short chains of 8-20 orthophosphate residues, which corresponds to data available in the literature [10]. The second sub-fraction (pH 4.5) contains higher polymers of polyP with n = 25-55, which is to say that the chains of these compounds contains even more orthophosphate residues than the salt-soluble polyP2 fraction. This once again provides support for the hypothesis that the chain length of various polyP fractions depends not only on the method of their fractionating, but also on the growth conditions and development phase of the yeast culture [12].
As seen from the data presented, the yeast growth on the phosphate-free medium (point B) is followed not only by a drop of polyP amount, but also by a considerable shortening of the polymer chain length in the polyP1, polyP3, and polyP4 fractions. During starvation only the polyP2 fraction demonstrated an increase in the chain length, which is associated with an assumption put forward long ago that there is a possibility of the transfer of low-molecular-mass fragments from high-polymer to lower-polymer fractions [10].Fig. 2. Dependence of the polyP chain length of different fractions on Pi content in the cultivation medium. Designations are the same as in Fig. 1 except for: 1a) polyP1 fraction obtained at pH 4.5, and 1b) polyP1 fraction obtained at pH 8.2; n) the number of orthophosphate residues in the polyP molecule.
A comparison of the data presented in Figs. 1a and 2 shows that an intensive polyP synthesis under the conditions of overcompensation (point C) does not actually lead to the increase of polyP chain length (polyP4 fraction) or is followed only by a minor increase (polyP1 and polyP3 fractions), or even by a decrease of the polymer degree (polyP2 fraction). It is impression that during the first 2 h of growth on the complete medium active polyP synthesis occurs at the level of fairly low-molecular-mass polyP (chain length for each fraction), or hand in hand with active synthesis, intensive polyP consumption occurs simultaneously. Of interest is the fact of a sharp rise in the polyP chain length observed during further yeast growth on the complete medium (point D) against the background of total absence (polyP3 fraction) or a minor increase (polyP4 fraction) of polyP accumulation.
The presence of several enzyme systems of polyP synthesis in microorganisms has been proved. Polyphosphate kinase (ATP:polyphosphate phosphotransferase, EC 2.7.4.1) catalyzes polyP synthesis at the expense of the terminal phosphate of ATP and has been isolated in highly purified state from a number of bacteria [18] and yeast [19]. More recently it has been found that polyphosphate kinase is localized in yeast mainly in vacuoles and catalyses not polyP synthesis, but rather the formation of ATP from ADP and polyP [20]. Finally, it has been shown that polyphosphate kinase preparation isolated from a yeast extract represents Ap4A phosphorylase (diadenosine-5´,5´´´-P1,P4-tetraphosphate alpha,beta-phosphorylase) [21]. However, this work does not contradict the presence of polyphosphate kinase in membranes or some membrane fractions.
PolyP synthesis at the expense of a high-energy phosphate from 1,3-diphosphoglyceric acid with the involvement of 1,3-diphosphoglycerate-polyP-phosphotransferase (EC 2.7.4.17) has been detected in a number of organisms. This enzyme was first revealed in a mutant of Neurospora crassa deficient in adenine synthesis, and then this enzyme was found in a diversity of bacteria [10, 22]. The enzyme is possibly localized in volutin granules and synthesizes a low-polymer polyP [10, 23]. However, this route of biosynthesis in yeast has not yet been revealed.
Finally, recent studies showed that biosynthesis of polyP localized at the cell surface of the yeast was coupled to the synthesis of mannoproteins, one of the main components of the cell wall, at the level of their lipid intermediate, dolichylpyrophosphatemannose [2, 6]. The content of this polyP may comprise 20-30% of the total amount of cell polyP depending on the conditions of cultivation. If it is remembered that almost the same polyP amount is localized in the vacuoles, where the polyP synthesis may proceed with polyphosphate kinase under certain conditions [20], it becomes clear that the problem of synthesis of just about half of the yeast polyP has been yet unknown.
Both exopolyphosphatases splitting off orthophosphate residues from the polymer chain end and endopolyphosphatases cleaving the polyP chain into separate fragments without producing orthophosphate are involved in polyP degradation. Exopolyphosphatases (polyphosphate phosphohydrolase, EC 2.6.1.11) localized in various structures of the yeast cell are substantially distinguished from each other by their properties [9]. Endopolyphosphatase (polyphosphate depolymerase, polyphosphate polyphosphohydrolase, EC 3.6.1.10), which has long been detected in the fungi and yeast [10], has been purified to homogeneity only recently, and its primary localization in the yeast vacuoles has been shown although its presence in other cell compartments is not excluded [24]. In N. crassa, at least, this enzyme is found in the periplasmic space of the cell surface and in the nuclei [25].
While the level of polyP in cells depends on the relationship between polyP synthesis and consumption, the change of a polymer chain length is primarily associated with the action of exo- and endopolyphosphatases. It has been shown that in yeast and fungi, unlike the bacteria [26], the Pi concentration in the medium has no effect on the exopolyphosphatase activity [7, 27, 28]. On the contrary, Pi and polyP content in the cell finely regulate endopolyphosphatase activity in fungi. For example, high Pi concentration in N. crassa at low polyP level stimulates the activity of this enzyme and, on the contrary, high polyP level leads to inhibition of this activity [29].
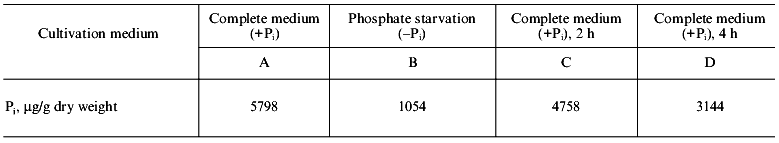
The data on Pi content in cells of S. cerevisiae under different conditions of the yeast cultivation are illustrated in the table. As seen, the lack of Pi in the medium within 7 h results in more than 5-fold drop in Pi in cells (point B). Then after the replacement of cells into a phosphate-containing medium, Pi concentration in cells is sharply increased (point C) and remains at a high level (point D) thereafter.
Pi content in cells of S. cerevisiae under different
cultivation conditions
Thus, as appears from the above, the endopolyphosphatase activity is likely to affect the variations in the chain length of different polyP fractions. While in the case of extended phosphate starvation (point B), polyP amount and the degree of their polymerization decrease in most fractions, a considerable polyP accumulation under overcompensation (point C) followed by endopolyphosphatase activation under these conditions leads to predominance of fairly low-polymer (unique for each fraction) polyP chains. High polyP content causes further inhibition of endopolyphosphatase and as a result polyP fractions at point D are represented by higher-molecular-mass polymers.
Finally, one more assumption must not be excluded, which could explain the elongation of the polyP chains in polyP3 and polyP4 fractions with their fixed content after replacement into Pi-containing medium. The assumption that exopolyphosphatases, which are able under certain conditions not to degrade but synthesize, just like an ATPase, may be involved in the polyP synthesis has already been put forward [2, 10]. It is not improbable that the endopolyphosphatase under certain conditions (lack of an active polymer synthesis de novo, high concentration of a substrate with particular chain length, etc.) will act as an enzyme joining together the low-molecular-mass fragments into high-molecular-mass polyP chains. If it is true, the polyP synthesis in the yeast may proceed in two stages: first the low-molecular-mass chains of these compounds are synthesized, and this is followed by high-molecular-mass polymers. To confirm this assumption further detailed investigations in this direction are required.
The authors would like to thank L. A. Sibeldina and I. N. Shchipanova for their help in 31P-NMR spectroscopy studies and fruitful discussion of the results obtained, and also A. V. Mudrik for her skillful technical assistance.
This work was supported by grants INCO-Copernicus PL971185, the Russian Foundation for Basic Research 99-04-48246, and also by the grant of Scientific Schools 96-15-98024.
REFERENCES
1. Kulaev, I. S., and Vagabov, V. M. (1983) Adv.
Microb. Physiol.,24, 83-171.
2. Vagabov, V. M. (1988) Biosynthesis of
Carbohydrate Components of Yeast Cell Wall [in Russian], NTsBI
Akad. Nauk SSSR, Pushchino.
3. Kornberg, A. (1995) J.
Bacteriol.,177, 491-496.
4. Vagabov, V. M., Chemodanova, O. V., and Kulaev, I.
S. (1990) Dokl. Akad. Nauk SSSR,313, 989-992.
5. Ivanov, A. Yu., Vagabov, V. M., Phomchenkov, V.
M., and Kulaev, I. S. (1996) Mikrobiologiya,65,
607-612.
6. Kulaev, I. S., Vagabov, V. M., and Shabalin, Yu.
A. (1987) in Phosphate Metabolism and Cellular Regulation in
Microorganisms (Torriani-Gorini, A., Rothman, F. G., Silver, S.,
Wright, A., and Yagil, E., eds.) American Society for Microbiology,
Washington, pp. 233-238.
7. Trilisenko, L. V., Vagabov, V. M., and Kulaev, I.
S. (1982) Biokhimiya, 47, 1963-1969.
8. Trilisenko, L. V., Vagabov, V. M., and Kulaev, I.
S. (1985) Dokl. Akad. Nauk SSSR,280, 763-765.
9. Kulaev, I. S., Andreeva, N. A., Lichko, L. P., and
Kulakovskaya, T. V. (1995) Biochemistry (Moscow), 60,
1061-1067.
10. Kulaev, I. S. (1975) Biochemistry of
High-Molecular Polyphosphates [in Russian], Moscow State
University, Moscow.
11. Schuddemat, J., de Boo, R., van Leeuwen, C. C.
M., and van den Broek, P. J. A. (1989) Biochim. Biophys.
Acta,1010, 191-198.
12. Vagabov, V. M., Trilisenko, L. V., Shchipanova,
T. N., Sibel'dina, L. A., and Kulaev, I. S. (1998)
Mikrobiologiya,67, 188-193.
13. Reader, V. (1927) Biochem. J.,21,
901-903.
14. Kulaev, I. S., Belozersky, A. N., Kritsky, M.
S., and Kokurina, N. A. (1960) Dokl. Akad. Nauk SSSR,130,
667-669.
15. Pilatus, U., Mayer, A., and Hildebrandt, A.
(1989) Arch. Biochem. Biophys.,275, 215-223.
16. Weil-Malherbe, H., and Green, R. M. (1951)
Biochem. J.,49, 286-292.
17. Dubois, M., Gilles, R. A., Hamilton, J. K.,
Rebers, P. A., and Smith, F. (1956) Anal. Chem.,28,
350-356.
18. Kornberg, A., Kornberg, S., and Simms, E. (1956)
Biochim. Biophys. Acta,20, 215-227.
19. Felter, S., and Stahl, A. J. C. (1973)
Biochemie,55, 245-249.
20. Shabalin, Yu. A., Vagabov, V. M., Tsiomenko, A.
B., Zemlyanuhina, O. A., and Kulaev, I. S. (1977) Biokhimiya,
42, 1642-1648.
21. Booth, J. W., and Guidotti, G. (1995) J.
Biol. Chem.,270, 19377-19382.
22. Kulaev, I. S., and Bobyk, M. A. (1971)
Biokhimiya,36, 426-429.
23. Konoshenko, G. I., Umnov, A. M., Bobyk, M. A.,
Mansurova, S. E., and Kulaev, I. S. (1973) in Problems of Regulation
of Metabolism in Microorganisms (Kulaev, I. S., ed.) [in Russian],
NTsBI Akad. Nauk SSSR, Pushchino, pp. 275-285.
24. Kumble, K. D., and Kornberg, A. (1996) J.
Biol. Chem.,271, 27146-27151.
25. Kulaev, I. S., Konoshenko, G. I., Chernyshova,
E. I., and Kritsky, M. S. (1972) Dokl. Akad. Nauk
SSSR,206, 233-235.
26. Nesmeyanova, M. A., Dmitriev, A. D., and Kulaev,
I. S. (1974) Mikrobiologiya,43, 227-233.
27. Felter, S., and Stahl, A. J. C. (1970) Bull.
Soc. Chim. Biol.,52, 75-78.
28. Umnov, A. M., Steblyak, A. G., Umnova, N. S.,
Mansurova, S. E., and Kulaev, I. S. (1975)
Mikrobiologiya,44, 414-421.
29. Kritsky, M. S., Chernishova, E. I., and Kulaev,
I. S. (1972) Biokhimiya,37, 983-990.