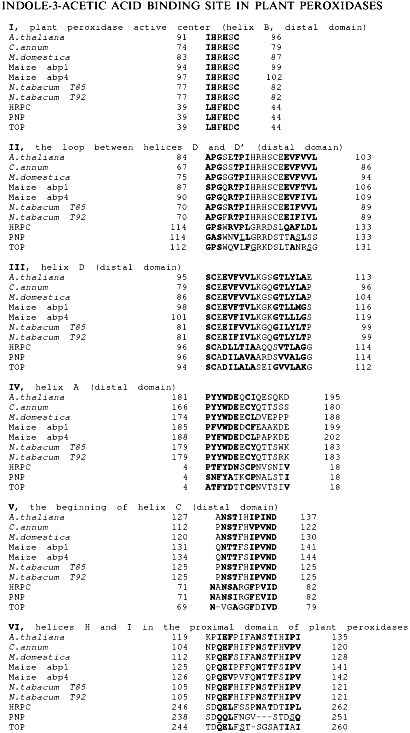
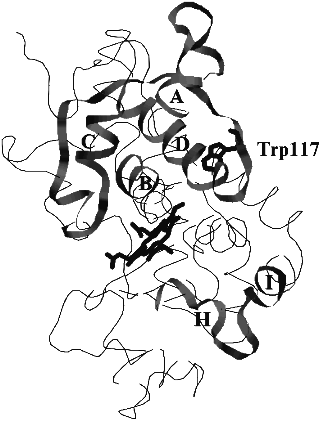
Evidence for Indole-3-Acetic Acid Binding Site in Plant Peroxidases. Structural Similarity between Peroxidases and Auxin-Binding Proteins
P. A. Savitsky1, A. M. Rojkova1, V. I. Tishkov1, I. V. Ouporov1, G. N. Rudenskaya2, and I. G. Gazaryan1*
1Department of Chemical Enzymology, School of Chemistry, Lomonosov Moscow State University, Moscow, 119899 Russia; fax: (095) 939-2742; E-mail: gazaryan@enzyme.chem.msu.su2Department of Chemistry of Natural Compounds, School of Chemistry, Lomonosov Moscow State University, Moscow, 119899 Russia
* To whom correspondence should be addressed.
Received February 17, 1998
Application of computer methods allowed us to demonstrate that plant peroxidases and auxin-binding proteins contain structurally similar fragments. The mapping of the fragments was done using a model structure of horseradish peroxidase. Five of six structurally similar fragments belong to the distal domain and form a subdomain in plant peroxidases that includes the distal heme-coordinating sequence, LHFHDC (amino acid residues 39-44 in horseradish peroxidase). The existence of a substrate-binding site for indole-3-acetic acid in the distal subdomain comprising helices A (whole), B (middle), C (beginning), and D (whole) and the loop between helices D and D' is discussed.
KEY WORDS: distal domain, active center, horseradish peroxidase, model structure, mapping, structurally similar fragments, substrate-binding site
Plant peroxidases (EC 1.11.1.7) catalyze oxidation of numerous electron donors with hydrogen peroxide in accordance with the following scheme [1]:
| E + H2O2 --> EI + H2O, | (1) |
| EI + S --> EII + P, | (2) |
| EII + S + H+ --> E + H2O + P, | (3) |
where E, EI, and EII are native enzyme and its compounds I and II, respectively; S and P are substrate and product of its one-electron oxidation.
The recent progress in refinement of crystal structures of these enzymes [2, 3] and their protein engineering [4, 5] allowed the mechanism of heterolytic cleavage of hydrogen peroxide at the enzyme active center to be elucidated. However, there is still no rational to predict the substrate specificity of an individual plant peroxidase because no substrate-binding site for a peculiar electron donor has been identified.
Plant peroxidases are known to be involved in metabolism of plant growth hormones--auxins, and indole-3-acetic acid (IAA) among them is the most powerful hormone responsible for plant growth and development. The mechanism of its oxidation by oxygen catalyzed by plant peroxidases remained obscure for many decades due to the complication of the enzymatic reaction with numerous radical processes. The recent studies on the mechanism of IAA oxidation catalyzed by plant peroxidases led to the conclusion on the necessary presence of a substrate-binding site for this physiological substrate [6-8].
We have proposed that plant peroxidases are highly specific IAA oxygenases and the reaction cycle is initiated via the formation of a ternary complex between the enzyme, IAA, and oxygen yielding IAA cation radical [6]:
| E3+ + IAA <--> [E-IAA] + O2 <--> [E-IAA-O2], | (4) |
| [E-IAA-O2] <--> E + IAA·+ + O2·-, | (5) |
where IAA·+ is IAA cation radical and O2·- is superoxide anion radical.
In acidic medium IAA cation radical decarboxylates yielding skatolyl radical (6). Skatolyl radicals react with molecular dioxygen yielding peroxy-radicals (7) and then skatole hydroperoxide via reaction (8):
| IAA·+ --> InCH2· + CO2, | (6) |
| InCH2· + O2 --> InCH2O2·, | (7) |
| InCH2O2· + IAA --> InCH2OOH + IAA·, | (8) |
where IAA· is indolyl radical and InCH2·, InCH2O2·, and InCH2OOH are skatole radical, peroxy-radical, and hydroperoxide, respectively.
We have just succeeded in identification and isolation of skatole hydroperoxide [7]. We showed it to be a reaction product degradable by catalase [7]. Thus, the inability of catalase to inhibit the oxygenase process provided indirect evidence for radical generation in a reaction alternative to the standard peroxidase cycle. Moreover, no compound I was detected in transient kinetic studies [6-8]. To explain this we have to propose that the enzyme molecule is able to bind simultaneously both skatole hydroperoxide and IAA, or in other words, that peroxidase contains a specific binding site for IAA which differs from the active center responsible for hydrogen peroxide and organic hydroperoxide cleavage.
The efficiency of oxidation of IAA and its analogs depends on the substrate structure [9, 10]. A true enzymatic character of IAA oxidation was first proposed by Ricard and Job [11]. These authors were the only ones who pointed out the reaction specificity and postulated IAA conversion directly into indole-3-aldehyde at the enzyme active center. Moreover, they supposed that IAA radical is kept at the enzyme active center and reacts with oxygen to yield compound II:
| E...IAA· + O2 --> EII + Ind-CH2O + CO2, | (9) |
where Ind-CH2O is a hypothetical intermediate, indole-3-epoxide.
This reaction is formally equivalent to a proposal on the productive interaction between the enzyme and peroxy-radicals. The unique character of reaction (9) is that peroxy radicals are consumed without damaging the enzyme active center while peroxy radicals derived from phenols under aerobic conditions are known to inactivate the enzyme [12]. The results of our recent studies on the reaction of IAA oxidation by molecular dioxygen in neutral medium in the presence of catalase [8] also indicated the existence of a reaction between the enzyme and skatole peroxy-radicals yielding compound II directly.
The goal of the present study was to find an IAA binding site in plant peroxidases. The data on cloning of auxin-binding proteins have been recently published and this allowed us to check if there is any homology between them and the known amino acid sequences of plant peroxidases. Application of computer methods shows that plant peroxidases contain fragments structurally similar to auxin-binding proteins. Moreover, these fragments were shown to organize the major part of the distal domain in plant peroxidases, and one of them directly corresponds to the sequence coordinating the heme at the distal site of the active center.
MATERIALS AND METHODS
The amino acid sequences of peanut [13], horseradish [14], and tobacco [15] peroxidases and seven auxin-binding proteins from Arabidopsis thaliana (GenBank Accession Z48451), Capsicum annum (GenBank Accession X69901), Malus domestica (GenBank Accession U77952), maize (GenBank Accession L08425, L08426), and Nicotiana tabacum (GenBank Accession X70902, X70903) were analyzed for structural similarity.
Structurally similar fragments were mapped using a horseradish peroxidase model structure created by I. V. Ouporov by means of homology modeling on the basis of peanut peroxidase crystal structure. The model is principally similar to the real structure of recombinant horseradish peroxidase whose coordinates were kindly provided by Dr. M. Gajhede (University of Copenhagen, Denmark) for the comparison. The distal domains in the model and the crystal structure are indistinguishable while the proximal domains are shifted by 1-1.5 Å.
Search for structural similarity was performed using the MACAW program, version 2.0.0, with the BLAST algorithm [16]. Structurally similar fragments were mapped on the model structure of horseradish peroxidase using RASMOL program, version 2.5.
RESULTS AND DISCUSSION
The search for structural similarity between heme-containing peroxidases and auxin-binding proteins first performed in this work was successful. Six structurally similar fragments presented in Fig. 1 were identified. Their mapping in the model structure of horseradish peroxidase which principally corresponds to the real structure of the enzyme is shown in Fig. 2. Unfortunately, there is no information concerning the structures of the sequenced auxin-binding proteins, and thus, the following discussion is based on the crystal structure of plant peroxidases.
Fig. 1. Amino acid sequences of structurally similar fragments of auxin-binding proteins and plant peroxidases. HRPC, horseradish peroxidase C; PNP, cationic peanut peroxidase; TOP, anionic tobacco peroxidase.
The first fragment, I, presented in Fig. 1, exactly corresponds to the sequence coordinating the heme from the distal site. This fragment belongs to helix B and is highly conserved through all heme-containing peroxidases. However, only classic plant peroxidases contain two histidine residues in this region, viz. 40 and 42. The sequences flanking this fragment in auxin-binding proteins are similar to the loop between helices D and D' in plant peroxidases (residues 114-133 in Fig. 1, II). Auxin-binding proteins also contain fragments structurally similar to helix D (residues 97-114 in Fig. 1, III). Two other structurally similar fragments correspond to the helix A (residues 4-18) and the beginning of helix C (residues 71-82) in the structure of peanut peroxidase (Fig. 1, IV and V). These five fragments presented in Fig. 1, I-V, form a specific subdomain (helices A, B, D-D', and C) in the distal domain of plant peroxidases.Fig. 2. The model structure of horseradish peroxidase. The structurally similar fragments corresponding to helices A, B, C, and D in the distal domain and H and I in the proximal domain, heme, and Trp-117 are shown.
Another similar fragment in the proximal domain of plant peroxidases (Fig. 1, VI) is unlikely to play any catalytic role, being apart from the active center and extensively glycosylated. It contains three oligosaccharide chains in the HRP molecule [3]. Moreover, it is the only one similar fragment in the peroxidase proximal domain compared to the presence of five structurally similar fragments in the distal domain.
Thus, we have now structural evidence for the presence of a special binding site for IAA in plant peroxidases. This conclusion is in agreement both with our proposal made on the basis of the mechanistic studies on IAA oxidation [7, 8] and with the results of the earlier work on auxin-binding protein isolation from A. thaliana using affinity chromatography, when both auxin-binding proteins and peroxidase were adsorbed on a IAA-modified sylochrom [17].
We think that the presence of Trp-117 (Fig. 2) in the subdomain organized by fragments structurally similar to those of auxin-binding proteins is principal for the enzyme catalytic activity. All plant peroxidases sequenced so far contain only one Trp residue whose position is highly conserved [18]. One can assume either an important catalytic role or a key structural role played by this single Trp residue in plant peroxidases.
Such a proposal is based on the known functions of tryptophan residues in cytochrome c peroxidase and lignin peroxidase. In the case of cytochrome c peroxidase Trp-191 adjacent to the porphyrin ring reduces porphyrin pi-cation radical and yields compound I with the protein-bound radical distinguishing this enzyme among other heme-containing peroxidases [19]. In the case of lignin peroxidase, both groups of crystallographers described a special modification of Trp-171 (a hydroxylated beta-carbon atom in the tryptophan residue) [20, 21]. However, the recombinant enzyme refolded from E. coli inclusion bodies exhibits no such modification. It appears only after the reaction cycle with hydrogen peroxide proceeds [22]. The replacement of this tryptophan residue with phenylalanine knocks out the enzyme activity toward its physiological substrate, veratryl alcohol, while the activities toward artificial electron donors are preserved unchanged [23]. Thus, there is an electron transport chain between the active center of lignin peroxidase and Trp-171 located ~16 Å apart.
In the crystal structure Trp-117 is 8-9 Å apart from N-atoms of the porphyrin ring and is in the zone of electron tunneling. It is likely that this tryptophan residue plays a catalytic role in the IAA-binding site distinct from the active center of plant peroxidases. The subdomain including structurally similar fragments in the distal domain of peroxidase is formed towards the surface of the distal domain, opposite to the entrance to the active center.
We think that the IAA-binding site could differ from the active center taking into account the analogous mechanism of action of prostaglandin H synthase, the enzyme catalyzing oxidation of arachidonic acid with molecular dioxygen in the presence of electron donor substrates. It has been shown that arachidonic acid is bound ~12 Å apart from the active center in a separate domain [24]. The reaction mechanism involves the formation of an intermediate hydroperoxide which reacts in accordance with the common peroxidase cycle [25]. However, compound I cannot be detected [25]. We propose that the distinct binding site for arachidonic acid allows the donor substrate and organic hydroperoxide (prostaglandin G2) to be bound on the enzyme simultaneously. In this case compound I oxidizes arachidonic acid at once and is converted into compound II due to the existence of an electron transport chain between heme and the substrate-binding site.
Our identification of skatole hydroperoxide as a key product of IAA enzymatic degradation [7] and the absence of compound I in the reaction [6-8] suggested similarities between the action of plant peroxidases and prostaglandin H synthase in the reaction of oxidation of their physiological substrates with molecular dioxygen. Taking into account the structural similarity between plant peroxidases and auxin-binding proteins discovered in this work, we consider that the distal domain must contain a substrate-binding site for IAA. Thus, we identify the domain directly related to the physiological substrate of plant peroxidases. Its site-specific mutagenesis will answer the question on the molecular determinants of the substrate specificity of this class of enzymes.
REFERENCES
1.Dunford, H. B. (1991) in Peroxidases in
Chemistry and Biology (Everse, J., Everse, K. E., and Grisham, M.
B., eds.) Vol. 2, CRC Press, Boca Raton, FL, pp. 1-23.
2.Schuller, D. J., Ban, N., van Huystee, R. B.,
McPherson, A., and Poulos, T. L. (1996) Structure, 4,
311-321.
3.Gajhede, M., Henriksen, A., Schuller, D. J.,
Poulos, T. L., and Smith, A. T. (1996) Plant Peroxidases:
Biochemistry and Physiology, Abst. IV Int. Symp., July 6-10, 1996,
Vienna, Austria, p. O2.
4.Newmyer, S. L., and Ortiz de Montellano, P. R.
(1996) J. Biol. Chem., 271, 14891-14896.
5.Rodriguez-Lopez, J. N., Smith, A. T., and
Thorneley, R. N. F. (1996) J. Biol. Chem., 271,
4023-4030.
6.Gazaryan, I. G., Lagrimini, L. M., Ashby, G. A.,
and Thorneley, R. N. F. (1996) Biochem. J., 313,
841-847.
7.Gazarian, I. G., Lagrimini, L. M., Mellon, F. A.,
Naldrett, M. J., Ashby, G. A., and Thorneley, R. N. F. (1998)
Biochem. J., in press.
8.Gazarian, I. G., and Lagrimini, L. M. (1998)
Biophys. Chem., in press.
9.Hinman, R. L., and Lang, J. (1965)
Biochemistry, 4, 144-158.
10.Kobayashi, S., Sugioka, K., Nakano, H., Nakano,
M., and Tero-Kubota, S. (1984) Biochemistry, 23,
4589-4597.
11.Ricard, J., and Job, D. (1974) Eur. J.
Biochem., 44, 359-371.
12.Ma, X., and Rokita, S. E. (1988) Biochem.
Biophys. Res. Commun., 157, 160-165.
13.Buffard, D., Breda, C., van Huystee, R. B.,
Asemota, O., Pierre, M., Dang Ha, D. B., and Esnault, R. (1990)
Proc. Natl. Acad. Sci. USA, 87, 8874-8878.
14.Welinder, K. (1979) Eur. J. Biochem.,
96, 483-502.
15.Lagrimini, L. M., Burkhart, W., Moyer, M., and
Rothstein, S. (1987) Proc. Natl. Acad. Sci. USA, 84,
7542-7546.
16.Altschul, S. F., Gish, W., Miller, W., Myers, E.
W., and Lipman, D. J. (1990) J. Mol. Biol., 215,
403-410.
17.Kartashova, E. P., Rudenskaya, G. N., Afifi, V.
M., Stepanova, V. M., and Gusev, M. V. (1985) Biol. Nauki,
12, 81-88.
18.Gazaryan, I. G., and Egorov, A. M. (1996)
Advances in Molecular and Cell Biology (Bittar, E. E.,
Danielsson, B., and Bulow, L., eds.) Vol. 15A, JAI Press Inc.,
Greenwich, Connecticut, pp. 59-68.
19.Erman, J. E., Vitello, L. B., Mauro, J. M., and
Kraut, J. (1989) Biochemistry, 28, 7992-7995.
20.Poulos, T. L., Edwards, S. L., Wariishi, H., and
Gold, M. H. (1993) J. Biol. Chem., 268, 4429-4440.
21.Piontek, K., Glumoff, T., and Winterhalter, K.
(1993) FEBS Lett., 315, 119-124.
22.Blodig, W., Choinowski, T., Winterhalter, K., and
Piontek, K. (1997) Forty-Sixth Harden Conference “Structure and
Mechanism of Oxidases and Related Systems” Programm and
Abstracts, Plymouth, UK, August 28-September 2, 1997, P4.
23.Doyle, W. A., Cottin, M., Veitch, N., and Smith,
A. T. (1997) Forty-Sixth Harden Conference “Structure and
Mechanism of Oxidases and Related Systems” Programm and
Abstracts, Plymouth, UK, August 28-September 2, 1997, 10.
24.Picot, D., Loll, P. J., and Garavito, R. M.
(1994) Nature, 367, 243-249.
25.Lambeir, A.-M., Markey, C. M., Dunford, H. B.,
and Marnett, L. J. (1990) J. Biol. Chem., 260,
14894-14896.