Telomerase Is an Unusual RNA-Containing Enzyme. A Review
S. S. Dokudovskaya,1 A. V. Petrov,1 O. A. Dontsova,1 and A. A. Bogdanov1,2
1School of Chemistry, Lomonosov Moscow State University, Moscow, 119899 Russia; fax: (095) 939-31-81; E-mail: dontsova@ribosome.genebee.msu.su2To whom correspondence should be addressed.
Submitted July 15, 1997.
Telomeres, the natural ends of linear eukaryotic chromosomes, are essential for protecting chromosomes from degradation and fusion. The synthesis of telomere DNA repeats in most eukaryotes is performed by a special enzyme, telomerase. Telomerase, a ribonucleoprotein enzyme, is a specialized reverse transcriptase utilizing its RNA moiety as a template for synthesis of telomeric DNA. Enzymatic properties and results of comparative analysis of telomerase RNA and protein structures from different eukaryotic systems are discussed in this review.
KEY WORDS: telomeres, telomerase, telomerase RNA, telomerase associated proteins.
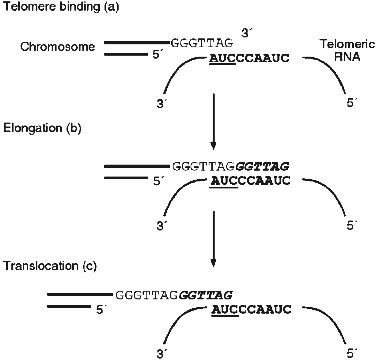
Eukaryotic chromosomes have on their ends special DNA--protein structures, telomeres. More then 50 years ago B. McClintock showed [1] that telomeres are essential for protection of chromosomes from degradation and fusion. Also, during the early development of molecular biology the problem concerning replication of linear chromosomes ends arose [2, 3]. DNA-polymerase is able synthesize only in the 5´-->3´ direction using RNA-primers, but conventional DNA polymerases cannot synthesize the extreme 5´-end of a blunt end DNA molecule. Thus, with every cell division chromosomes become shorter. This fact suggested the proposal of an interesting hypothesis about a new function of chromosomes as a mitotic clock which count out the lifetime of the cell [2]. In the majority of eukaryotes telomeric DNA consists of several repeats each with the length of 6-25 base pairs. One of the DNA strands contains mainly G-residues and at the very end of the G-strand there is a single-stranded segment consisting of several telomeric repeats (G-overhang). The mechanism of synthesis of the DNA-repeats themselves became clear only in 1985 when Greider and Blackburn [4] discovered an unusual enzyme, telomerase. It was shown that in the presence of dTTP and [alpha-32P]dGTP, an extract of Tetrahymena thermophila cells was able to elongate the oligonucleotide d(TTGGGG)4, which was used as an analog of telomeric G-strand. Later it was found that telomerase contains an RNA component [5]. Moreover, for enzymatic activity telomerase requires both the RNA and protein components. But the most interesting feature of telomerase is the fact that telomerase RNA contains a template for synthesis of telomeres [6]. Thus, telomerase is an unusual reverse transcriptase. The mechanism of elongation of the chromosome ends is schematically represented in Fig. 1; at the beginning the telomeric G-strand associates with the RNA template and afterwards the RNA-dependent synthesis of telomeric DNA and translocation takes place. The complementary chain is synthesized by DNA-polymerase.
In the middle of the 1990s it was shown that in accordance with the hypothesis proposed previously [2], telomerase as an enzyme compensating shortening of chromosomes is one of the characteristics of a cancer cell [7-10]. It was found that in cancer cells telomeres are rather short and very stable and the telomerase activity is very high [11-14]. At the same time, in almost all human somatic cells telomerase activity is not detected and telomeric DNA, very long at birth (12-15 kb), shortens with age. This fact explains the extremely high interest in telomerase and stimulated more intensive investigation of this enzyme.Fig. 1. Scheme of the chromosome end elongation mechanism.
Detection of Telomerase Activity in Cell Extracts. The study of telomerase has been mainly performed using the ciliate Tetrahymena thermophila because a single cell of this organism has more then 40,000 telomeres. So far the only purified enzyme was isolated from Tetrahymena thermophila [15]. When we discuss the properties of telomerase from other organisms, we will be describing the properties of cell extracts possessing telomerase activity.
To obtain cell extracts with telomerase activity, cells are usually harvested up to the S-phase of the cell cycle. It is known that during S-phase the synthesis of DNA is the most intensive. There are some data indicating that the maximum activity of telomerase is also detected during S-phase [16-18]. After degradation of the cell membranes an extract is purified using gel filtration, affinity chromatography, and centrifugation [15, 19, 20]. It is quite possible that during purification some telomerase inhibitors are eliminated, but so far there has been no attempt to find inhibitors in fractions without telomerase activity.
There are now two main approaches for the detection of telomerase activity--direct assay and assay based on PCR technique (the TRAP assay). In the first case, in addition to cell extract, the reaction mixture contains a telomeric primer (an oligonucleotide consists of one or several telomeric repeats), buffer with Mg2+ and dNTP mixture (one of which is radioactive, usually [alpha-32P]dGTP) [4-6]. In the second case, the DNA obtained after elongation of a telomeric primer serves as a template for the synthesis of the complementary DNA-chain using the PCR technique. The modification of the TRAP assay allows the elongation and amplification reaction in the same test tube [9]. The products of the elongation reaction are analyzed on a sequencing gel and a banding pattern with periodicity is seen on a film. Both methods have advantages and disadvantages. A TRAP assay is more then 104 times more sensitive than direct assay, and the development of this method allows detection of telomerase activity in different mammalian tumors [9]. At the same time, the TRAP assay produces many artifact products and to confirm the presence of telomerase activity it is worth performing a direct assay.
Enzymatic Properties of Telomerases
Processivity. The ability of telomerases for processive elongation of DNA-primers in vitro and telomeres in vivo depends on both the source of telomerase and the reaction conditions. In most cases telomerase is not processive in vitro and elongates a DNA primer by not more than one repeat1. Telomerases from Saccharomyces cerevisiae [19], Xenopus laevis [21], and mouse [22] are also not processive in vitro. However, the telomerase from another yeast strain (Saccharomyces castelli) has moderate processivity and adds 5-8 tandem repeats to the DNA-primer [19]. It is interesting that processivity of Xenopus laevis, rat, and mouse telomerases can be enhanced by increasing the dGTP concentration [23]. Human telomerase generates many tandem telomere repeats in a processive manner in vitro [22].
1For telomerases the meaning of the term "processivity" differs from that of DNA- and RNA-polymerases or reverse transcriptases. For example the processivity of DNA-polymerase is defined as the number of nucleotides added by the enzyme to a growing chain between two subsequent acts of association-dissociation. Telomerase is considered as a processive enzyme if it elongates DNA-primer (or chromosomes) by several telomeric repeats. In this case one does not take into account whether telomerase dissociates from the template only once or there are many acts of association-dissociation. Telomerase is considered as a nonprocessive enzyme if it adds only one telomeric repeat to a primer or a chromosome end.
The processivity of Tetrahymena thermophila telomerase has been studied in much more detail than that of enzymes isolated from other organisms. It was shown that the enzyme is nonprocessive in vivo [24, 25] (as well as S. cerevisiae telomerase [26]). However, in vitro telomerase can be as processive as nonprocessive depending on the reaction conditions. When it uses telomeric primers with length more than 10 nucleotides long, Tetrahymena thermophila telomerase shows a high processivity which does not depend on amount of the primer and does not change with increasing the primer length [27]. The size of the nascent DNA can reach the length of about 520 nucleotide residues. However, primers shorter than 10 nucleotides long are extended by addition of only one repeat [28]. For those primers processivity can be improved by increasing the concentration of oligonucleotide (>5 µM). These facts suggest that telomerase has at least two binding sites for a primer: one site is the template region on the telomerase RNA and the other site is the "anchor site". Binding to the "anchor site" prevents a product from dissociation and allows telomerase to act as a processive enzyme. There are two current models for anchor-site binding of a primer. According to the first model, the 5´-end of a primer remains bound to the anchor site and the growing product is "looped out". The second model suggests that the growing chain slides through the anchor site during processive elongation [29-31]. The latter model is based on the formerly proposed mechanism for processive elongation of RNA chains by RNA polymerases [32, 33].
Substrate Specificity. As mention above, telomerase has a high affinity to telomeric primers (Km = 5-10 nM) and elongates them with high efficiency. Earlier it was considered that the enzyme recognizes specific structures formed by G-rich oligonucleotides [4], but this hypothesis was not confirmed [34]. Then it was assumed that guanine bases themselves in the G-rich part of the primer are necessary for the recognition by the enzyme and subsequent elongation of the product. It was shown that nontelomeric primers can be extended if they have a telomeric repeat on their 3´- or 5´-ends [35] or even G-rich nontelomeric 5´-end proximal region [36]. Primers lacking any telomeric sequences were not elongated by telomerase even at a very high concentration (>10 µM). However, it was recently shown [37] that under certain conditions Tetrahymena thermophila telomerase elongates with high efficiency oligonucleotides with random sequence which were added to the system in nanomolar concentrations. Elongation of completely nontelomeric primers occurs if the Mg2+ concentration is not less than 1.25 mM and potassium and sodium ions are absent. In the same mode as in vivo, at the first stage telomerase adds the telomeric block dGGGGT to the primer 3´-end and the efficiency of the elongation depends on the sequence of both 3´- and 5´-ends of the oligonucleotides. The elongation of the nontelomeric primers by Euplotes crassus telomerase starts with the same sequence [38]. Moreover, Tetrahymena thermophila telomerase elongates nontelomeric primers only if they are not shorter than 30 nucleotides long, and double-stranded DNA-primers can serve as substrates for elongation if they have G-overhang more than 20 nucleotides long. These data serve as additional evidence for the existence of an "anchor site" on telomerase.
Tetrahymena thermophila telomerase can use hybrid DNA--RNA primers as substrates in an in vitro system [39]. It has been shown that the presence of three ribonucleotides at the 3´-end of a primer decreased the yield of the product but increased the affinity of a hybrid primer to telomerase. Since RNA--RNA duplexes are more stable than RNA--DNA duplexes, the presence of a RNA-block in a primer will increase its binding and inhibit its elongation.
Telomerase catalyzes the incorporation of deoxy- and dideoxyribonucleoside triphosphates into a polynucleotide chain with the same efficiency (Km = 1-10 µM) [27] although in the second case the termination of DNA synthesis occurs. However, in vitro Tetrahymena thermophila telomerase uses as substrates also rGTP and rTTP and processive synthesis occurs under the concentration of 10-100 µM of rNTPs but the yield of the product is less than in the case of dNTPs [39].
Thus, all the telomerase properties mentioned above together with data on the primary structure of telomerase protein p95 (see section "Telomeric Proteins" of this review) might suggest that RNA-dependent RNA-polymerases were ancestors of telomerases during evolution.
Exonuclease Activity. In addition to the ability to elongate primers, telomerases possess an exonuclease activity which allows them to hydrolyze both primers and products of enzyme synthesis. For the first time nucleolytic activity was found for Tetrahymena thermophila telomerase [28] which was able to remove the extreme 3´-end nucleotide residue from primers (GGGTTX)3 with X = A, G, T, C. However, other primers such as (GGGGTT)3 were not subjected to enzymatic hydrolysis. After reconstitution of telomerase activity hydrolytic activity was also restored (see section "Reconstitution of Telomerase Activity"). Mutant telomerase (A43U) with the template region UAACCCCAA removed the extreme 3´-G from d(GGGTTG)3 and after that DNA synthesis started from dA [40]. Thus, 5´-end of a template is a component of hydrolytic center of Tetrahymena thermophila telomerase.
Hydrolytic activity was also found for Euplotes crassus telomerase [38], but in contrast to Tetrahymena telomerase the enzyme is able to eliminate several nucleotides in primers which have a random sequence on their 3´-ends and telomeric DNA-block at the 5´-end. Telomerase eliminated all non-telomeric nucleotides and DNA synthesis always started from dG.
Telomerase from S. cerevisiae can hydrolyze up to 8 internucleotide bonds from the 3´-end [19]. However, in contrast to the Euplotes crassus enzyme the yeast enzyme can hydrolyze telomeric product or primer. At present there are no data concerning the hydrolytic activity of telomerases from other organisms.
It is hard to say now whether telomerase hydrolytic activity provides the accuracy of telomeric DNA synthesis since the fidelity of this process is not very high. In the case of Tetrahymena thermophila telomerase hydrolysis occurs at sites of natural pausing in elongation, which happen after addition of dGTP to the 5´-end C of a template region. This property of telomerase points out their similarity with known DNA-dependent RNA-polymerases [41, 42].
Structure and Function of Telomerase RNAs
Telomerase RNAs are much better studied than telomerase proteins. Synthesis of telomerase RNAs in different organisms is carried out by different RNA-polymerases. Telomerase RNAs of lower eukaryotes are synthesized by RNA-polymerase II [6, 24, 43, 44] and mammalian telomerase RNAs (mouse and human) are synthesized by RNA-polymerase III [45]. Moreover, yeast telomerase RNA is thought to be synthesized by both polymerases [46] and this obvious contradiction must be solved. In all organisms studied the gene of telomerase RNA exists in one copy per cell.
More than 30 genes of telomerase RNA from ciliates [47], yeast S. cerevisiae [46] and Kluyveromyces lactis [48], mouse [49], and human (HeLa) [45] have been cloned. The RNAs encoded by these genes differ very much in their primary structure and RNA sequences are not conservative not only in evolutionary distant organisms but also is closely related ones. Nevertheless, secondary structure of ciliate RNAs is very conservative [50]. It is likely that common features which are responsible for basic functions of telomerase can be revealed only on the level of tertiary structure.
Primary Structure of Telomerase RNAs. As mentioned above, the primary structure of different telomerase RNAs is not conservative. There is a difference not only in nucleotide sequence but also in the length of RNAs which increases from lower to high eukaryotes. In ciliates the length of RNAs vary from 147 to 209 nucleotides [51], and human and mouse have telomerase RNAs about 450 nucleotides long [45, 49]. However, yeast RNA is surprisingly long; it consists of 1301 nucleotide residues [46]. It is not clear yet why yeast telomerase has such a large RNA and whether all the nucleotides are necessary for telomerase functional activity. Such long RNAs may be a specific feature of yeast. For example small nuclear RNA U2 of S. cerevisiae (1175 nucleotide long) is almost 1000 nucleotides longer than its RNA analogs in mammals [52]. It is interesting that in most telomerase RNAs, the template is situated at a distance about 50 nucleotides from the 5´-end and only in yeast is it located at a distance of 450 nucleotides from the 5´-end.
Nucleotide sequences of telomerase RNAs differ very much even in related organisms. For instance, the sequences of genes of telomerase RNA from Tetrahymena thermophila and Tetrahymena hyperangularus share only 72% identity [53]. However, the sequence of ribosomal RNAs genes have 98% identical residues [50].
The gene of human telomerase RNA was used as a probe for searching for the analogous gene in the mouse [49]. It was found that sequence similarity in RNAs of the two organisms was only 65%, which is much less than for other small RNAs in these organisms [54]. Moreover, in spite of the fact that both RNAs encode the same telomeric repeat, the template region in mouse RNA consists of 8 nucleotides whereas in human it consists of 11 nucleotides.
Secondary Structure of Telomerase RNAs. The secondary structure of ciliate telomerase RNAs and some related lower eukaryote RNAs is studied in many details. Comparison of the sequence of the telomerase RNA from five Tetrahymena species and the related ciliate Glaucoma chattoni suggest a conserved secondary structure for these RNAs [50]. The corresponding model is based on covariation analysis of residues in putative helical regions of the aligned sequences. Later the phylogenetic model was proved by enzymatic and chemical probing in solutions of naked telomerase RNAs synthesized by T7 RNA-polymerase [55].
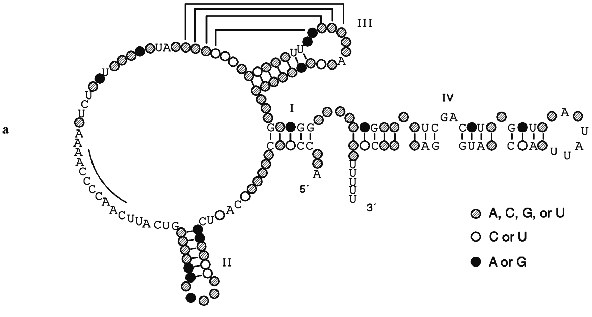
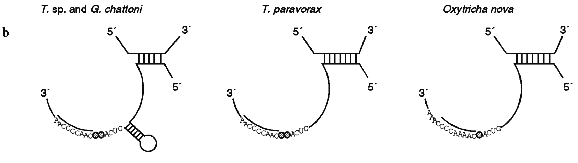
The secondary structure of Tetrahymena telomerase RNAs is shown in Fig. 2. The main structural elements of this model are four helical regions and an unstructured domain which includes the RNA template for synthesis of telomeres. Helix I consists of 5 base pairs in most ciliate telomerase RNAs although its length in Oxytricha nova is 9 bp. Helix II is absent in some evolutionarily earlier organisms such as Tetrahymena paravorax and Oxytricha nova (Fig. 2b). It was suggested that helix II is perhaps a non-essential functional element [55]. Deletion of this helix could confirm or disproof this suggestion, but so far this experiment has not been done.
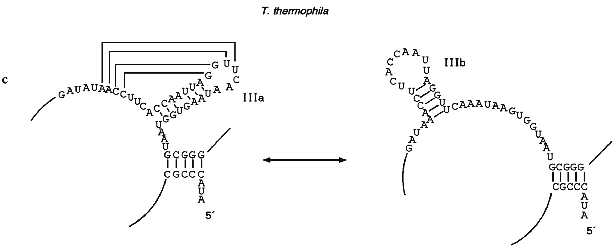
Helix III is absolutely conservative in all ciliates and forms a pseudoknot due to formation of Watson--Crick base pairs on interaction between four bases in the loop of helix III and the 5´ single-stranded region contiguous to this helix. The distance between helix III and the single-stranded region which is involved in the formation of the pseudoknot is usually 2-6 nucleotides although in T. paravorax the distance is very short, i.e., only one nucleotide. In T. paravorax the pseudoknot is formed by the interaction of only 3 nucleotides. There are some data indicating that pseudoknots similar to pseudoknots discovered in telomerase RNAs can be in equilibrium with alternative helices [47]. Enzymatic and chemical probing of telomerase RNAs from Tetrahymena thermophila and Glaucoma chattoni [55] in vitro showed that helix III together with the neighboring region can be transformed to an alternative hairpin IIIb (Fig. 2c). It was suggested that a flexible pseudoknot, formed by helix III, can influence the translocation process during telomerase-dependent synthesis of telomeres [47].Fig. 2. The secondary structures of Tetrahymena telomerase RNAs.
Helix IV is the longest double-stranded region of telomerase RNA but it is partitioned into two almost equal parts by a GA bulge. This bulge is absolutely conservative and was found in almost all telomerase RNAs of lower eukaryotes. Moreover, in the loop of helix IV there is the four nucleotide sequence 5´-UAUU-3´ which is also highly conservative. Helix IV most probably interacts with helix I. But the most important fact is that the GA bulge can introduce an appreciable kink of about 60°. It might be that the conserved bulge in helix IV distorts the helical trajectory in a structurally conserved manner and may serve as a recognition motif for protein binding.
The single-stranded region between helices II and III plays the most important functional role, being a template for synthesis of telomeric DNA. Although all templates are different, at the 5´-region of the template there is a third highly conservative sequence (5´-GUCA-3´). According to data of phylogenetic and computer analysis, the region of RNA close to the 3´-end of the template must be single-stranded. However, chemical and enzymatic probing has shown that this region is rather structured. E. Blackburn suggested [55] that this region together with the flexible pseudoknot formed by helix III may be part of the enzymatic center of telomerase RNA.
Multiple Functions of the Template Region. The main function of this region is to be the template for synthesis of telomeric DNA. Any changes in the template sequence produce complementary changes in telomeres of ciliates, yeast, mouse, and human [24-26, 40, 45, 46, 48, 49]. Some of these mutation do not influence the activity of telomerase but others can lead to telomere shortening, senescence, and cell death.
The template of Tetrahymena thermophila telomerase RNA is the best studied. It was shown that 9 nucleotides from position 43 to 51 (5´-CAACCCCAA-3´) can serve as a template [6]. Thus, the template is 1.5 times longer than a telomeric repeat synthesized by telomerase. Nevertheless, in vitro and in vivo site-directed mutagenesis showed [56] that only 7 nucleotides (43-49) are templating and the enzyme can use as a template either nucleotides 43-48 or 44-49 [57]. Mutations in positions 43 and 48 results in premature dissociation of a primer after copying 45 nucleotides. Moreover, changing C to U at position 48 leads to incorrect pairing of neighboring nucleotides 46 and 47. It is interesting that the insertion of one C-residue between nucleotides 49 and 50 leads to additional incorporation to telomeric DNA from 1 to 30 G-residues both in vivo [25] and in vitro [56]. Nucleotides 50-51 serve only for binding of DNA-primer of the 3´-end of chromosome. It was found [57] that in vitro mutation A50G and A50G-A51G result in an elongation of a primer only by one telomeric repeat. Surprisingly, the same mutation in vivo didn't change the phenotype and this can be and additional proof of the nonprocessive mode of elongation in vivo of Tetrahymena telomerase. Nevertheless, Watson--Crick base pair at position 50 is absolutely necessary for successful elongation in vitro.
The study of yeast telomerase from S. cerevisiae confirms that the template region is a component of an active center of an enzyme [26]. Insertion of GUG in different positions of a template influence binding of a primer and in diploid cause senescence and cell death. Very interesting effects were obtained after mutation of the template region of telomerase RNA from K. lactis [48]. Some changes resulted in uncontrolled elongation of telomeres which finally became 100 times longer than telomeres in wild type cells. Several mutations displayed their effects immediately, whereas others behave like "genetic time bombs" causing elongation only after a latent period of hundreds of generations.
Thus, it is clear that in addition to its main function--to be a template for synthesis of telomeric DNA--telomerase RNA takes part in catalytic activities of telomerase. Nontemplate regions of telomerase RNA are still not sufficiently studied from the functional point of view. One experimental evidence for the importance of nontemplating parts of telomerase RNA was found recently in Blackburn's group [58]. Telomerase RNA from ciliate Glaucoma chattoni was expressed in Tetrahymena thermophila cells. The template region base sequence is identical in two RNAs, but elsewhere their sequences differ by 49%. This hybrid telomerase enzyme was enzymatically active but added only short stretches of telomeric repeats in vivo and in vitro. This new enzyme also had strong, aberrant DNA cleavage activity in vitro.
Reconstitution of Telomerase Activity. To study the mechanism of telomerase functioning, it was necessary to develop a technique which would allowed reconstitution of active enzyme from RNA and proteins in vitro. Reconstitution conditions were first reported for Tetrahymena thermophila telomerase [40]. Telomerase RNA isolated from purified cell extract was digested with micrococcal nuclease followed by inactivation of the enzyme with EDTA. Then telomerase RNA synthesized by T7 transcription in vitro was added to protein extract in the presence of Mg2+. A new telomerase extract was able to elongate telomeric DNA-primer although the DNA-synthesized activity restored only by 10%. Moreover, this level of activity was achieved only when the RNA transcript was added in 1000-fold excess. One can suggest several explanations of these data: "incorrect" conformation of RNA transcript, loss of modified bases which can be important for telomerase functioning, incomplete inhibition of micrococcal nuclease, partial inactivation of telomerase proteins and/or protein factors acting as activators of telomerase. Nuclease treatment could be avoided if more gentle methods were applied for eliminating the wild telomerase RNA. The last problem will be overcome if superproducing strains of Tetrahymena telomerase proteins (which were sequenced recently [15]) will be obtain. However, protein cloning and expression is a rather complicated task since the genetic code in Tetrahymena is different from the standard one and needs to be changed. From the other side, it is hard to predict how the absence of modified residues in synthetic telomerase RNA will influence the activity of the enzyme. For example, it is known that it is not possible to reconstitute an active large ribosomal subunit from purified proteins and unmodified 23S RNA-transcript [59, 60]. However, reconstitution of the small ribosomal subunit from proteins and T7 transcript of 16S RNA give rise to an active ribosomes and this technique has been widely applied in studies of the structure and function ribosomes [61].
The same approach was used for reconstitution of active human telomerase [62]. In this case it was possible to obtain telomerase activity not only with full-length telomerase RNA transcript but also with shortened RNA-fragments. It was found that the functional region on telomerase RNA is situated between nucleotides 1-203. Moreover, it was shown that nucleotides 1-44 situated upstream from the template (45-56) are not necessary for enzymatic activity. However, mutation of full-length RNA in the region 170-199 completely inactivates the enzyme. Perhaps, this region is involved in the interaction with telomerase proteins. Thus, functionally important activity of human telomerase RNA is concentrated into a 203 nucleotide segment which is equal in length to ciliate telomerase RNAs. However, for ciliate RNAs the minimal RNA fragment necessary for enzymatic activity has still not been determined. Nevertheless, it would not be surprising if telomerase activity in ciliates can be restored only for full-length RNAs, but not for its fragments. It will also be interesting to compare the secondary structure of Tetrahymena RNAs with the secondary structure of telomerase RNAs from high eukaryotes. It might be expected that some conservative elements which play an essential role in telomerase functioning are located in their 5´-end regions.
Proteins of the Telomerase Complex
Tetrahymena thermophila was the first organism where protein components of purified telomerase complex (with molecular mass about 230 kD) were identified and sequenced [15]. It was shown that Tetrahymena thermophila telomerase consists of an essential 159 nucleotide RNA moiety (54 kD) and two polypeptide subunits, p80 and p95. The stoichiometry of p80/p95/telomerase RNA was estimated to be 1:1:1. After RNase treatment the two proteins remained associated.
Protein p80 binds to telomerase RNA with high affinity due to recognition by the protein of specific elements of secondary structure of telomerase RNA, for example the structural motif formed by helices IV and I (see above). Protein p95 interacts with the telomeric primer. Cross-linking experiments confirmed that p95 has a unique site for primer binding, the "anchor site". DNA binding with the "anchor site" does not required the presence of functional template and even telomerase RNA itself.
In spite of the fact that the sequences of p80 and p95 are completely different, they bear some resemblance to several RNA-polymerase sequences. Thus, p95 has a common sequence motif with the active center region of virus RNA-dependent RNA-polymerases. p95 also has a motif analogous to a structural motif of class II tRNA-synthetase, which being coordinated with Mg2+ by amino acid residues outside the motif, serves to position a reactive nucleotide triphosphate at the synthetase active site.
It is interesting that p80 has a putative metal-binding motif CxxC(27)CxxC which is similar to zinc-binding motifs of transcriptional factors TFIIS and TFIIE. It is known that TFIIE affects initiation of transcription, promoting movement of RNA polymerase from a stable open promoter complex. TFIIS is also necessary for RNA-polymerase movement along a DNA-chain during elongation. In addition, this factor stimulates nucleolytic cleavage of the RNA product. It is interesting that the nucleolytic cleavage reaction of RNA polymerases has many similarities with the nucleolytic cleavage reaction catalyzed by Tetrahymena thermophila telomerase [28].
Isolation of a functional telomerase complex with a protein moiety consisting of two polypeptide chains with molecular masses 123 and 43 kD was also reported for Euplotes aediculatus [20]. In contrast to Tetrahymena thermophila, the "anchor site" of Euplotes aediculatus telomerase is formed by protein p123 together with telomerase RNA [31]. Moreover, p123 has structural motifs similar to conservative motifs of reverse transcriptases [63].
In the case of yeast, data about telomerase proteins are rather contradictory. Genetic selection methods revealed the gene est1 [64] which encodes protein EST1 with molecular mass 77 kD. Some authors believe that EST1 is a component of yeast telomerase as far as deletion of the est1 gene leads to telomere shortening [64-66] and EST1, with epitopes fused to its N-terminus, is co-precipitated together with antibodies to yeast telomerase RNA [67, 68]. In contrast, in a haploid strain where more than 80% of the est1 gene was replaced by an another sequence, telomerase activity in vitro did not differ from telomerase activity in the wild-type strain [19]. Most probably protein EST1 is not a component of telomerase itself. It was shown [69] that in vitro EST1 interacts with telomeric primer and exhibits properties of SSB-proteins. Thus, EST1 might be one of the telomeric proteins although comparison of sequences of EST1 and RAP1 (yeast telomeric protein) did not reveal any similarity. There is an interesting hypothesis suggesting that EST1 takes part in biosynthesis of nucleotide precursors. The dependence of telomere metabolism on nucleotide biosynthesis was supported by the fact that two mutations which affect thymidine biosynthesis also change the length of telomeres [64]. Although the precise function of EST1 remains unclear, the data mentioned above suggest that this protein takes part in regulation of telomere synthesis.
Recently three more proteins of the EST family were found [70]. Deletion of genes encoding these proteins results in telomere shortening. One of these proteins--EST2--has homology with telomerase protein p123 from Euplotes aediculatus and like p123 has structural motif analogous to a highly conservative structural element from the active site of reverse transcriptases [63]. Thus, EST2 is a protein component of yeast telomerase.
Comparative analysis is now the most effective way of searching for new protein components of telomerase. For example, database mammalian gene homologs to the gene of Tetrahymena thermophila protein p80 were found and followed by identification of rat telomerase protein TLP1 [71] and human protein TP1 [72]. Both proteins are very large, 240 kD. It is interesting that in vivo rat protein TLP1 (p240) undergoes processing which transforms it to a shortened protein, p230. In telomerase-negative kidney, p240 was the predominant form; p230 was more abundant in the telomerase-positive testis. The liver, which had a moderate level of telomerase activity, displayed an intermediate pattern, with similar levels of p240 and p230. These results suggest that the modified p230 is associated with the active telomerase, whereas p240 precursor is associated with an inactive form. TLP1 has a potential ATP/GTP-binding domain and WD repeats. WD proteins contain highly conserved repeating units usually ending with Trp-Asp (WD). They are found in all eukaryotes. They regulate cellular functions such as cell division, cell-fate determination, gene transcription, transmembrane signaling, mRNA modification, and vesicle fusion [73]. WD domains of TLP1 might be a part of the catalytic center of telomerase or they could have some regulatory function. On the other hand, since the part of the polypeptide chain of TLP1 exhibits homology to p80 of Tetrahymena thermophila, one can suggest that TLP1 could have functions similar to p80.
Protein TP1 [72]--the human analog of p80 from Tetrahymena thermophila--also has a potential ATP/GTP motif and WD repeats. Using a three-hybrid screen together with immune coprecipitation the direct interaction of TP1 and telomerase RNA was shown. It is interesting that the N-terminus of TP1 homologs to p80 are sufficient for binding of human telomerase RNA in vivo. This may suggest the existence of some highly conservative element of telomerase RNA secondary (tertiary) structure in these evolutionarily distant organisms.
Activators and Inhibitors of Telomerase
The search for activators and inhibitors of telomerase became a very important task since the correlation between telomerase activity, senescence, and oncogenesis was established. Telomerase activity has been detected in many mammalian tumor tissues [9], and it is probably critical for sustained tumor proliferation [74], although this hypothesis remains in dispute [75, 76]. Thus, telomerase inhibition in cancer cells might be a viable strategy for suppression of tumor growth. On the other hand, activation of telomerase is linked to cell immortalization [8, 29].
An intensive search for both natural and synthetic inhibitors of telomerase is now being done. Thus, it was found that expression of the human papilloma virus type 16 E6 protein activates telomerase in early-passage human keratinocytes and mammary epithelial cells [77]. However, this is the only natural activator found and all other examples are synthetic inhibitors. There are now two main strategies in the search for telomerase inhibitors: expression of antisense DNA complementary to telomerase RNA and studies of the effects of known inhibitors of retroviral reverse transcriptases.
As expected, oligonucleotides complementary to template of telomerase RNA can inhibit telomerase [15] but this inhibition requires that the oligonucleotides be relatively long (18-40 nucleotides) and present at high concentration in order to compensate for hydrolysis with nucleases [43]. To overcome this problem, it has been suggested to use as inhibitors modified oligonucleotides, e.g., peptide nucleic acids and thiophosphate group-containing oligomers [78]. These oligonucleotides afford greater resistance to degradation by proteases or nucleases. It was shown that these oligonucleotides specifically interact with template of telomerase RNA and effectively inhibit telomerase activity in vitro. However, the application of such oligonucleotides is restricted very much in crude cell extracts or in vivo due to the main limitation of "antisence technology"--difficulties with transportation of oligonucleotides through membranes to the cell nucleus.
Because telomerase possesses similarity to viral reverse transcriptases, some inhibitors of these enzymes such as dideoxyguanosine (ddG) and azidothymidine (AZT) were tested as potential telomerase inhibitors [79, 80]. It was shown that ddG caused reproducible, progressive telomere shortening after which the telomere stabilizes and remains short. The prolonged passaging in ddG did not cause any observable effects on cell population doubling rates or morphology [79]. AZT caused progressive telomere shortening in some but not all T- and B-cell cultures. It is interesting that prolonged passages in dideoxyadenosine, dideoxyinosine, and didehydrothymidine did not cause reproducible telomere shortening or decreased cell growth or viability.
Perspectives
Telomerase activity was found by Blackburn and Greider 12 years ago [4]. Since that time much has been learned about telomerase properties and structure. However, the main stage in telomerase research is starting now. One can predict that further investigation of telomerase will go in three main directions.
First, the study of structure and function of the enzyme will be prolonged. Although telomerase RNAs are studied rather well, we still do not have any experimental data about their tertiary structure. Moreover, even secondary structure of telomerase RNAs from higher eukaryotes is still not determined. On the other hand, only two years ago the first data on telomerase protein components became available and the complete set of proteins is known only for Tetrahymena thermophila. It appears that in the near future some progress in identification of telomerase protein components will be achieved. Thus, an opportunity to study RNA--protein interactions in telomerase will be open that will surely help to identify not only enzyme structure but the mechanism of telomerase functioning.
Second, the understanding of the role of telomerase in regulation of telomere synthesis is of great interest. It has become more evident that telomere maintenance depends on many factors such as interaction with telomerase, telomeric proteins, and some still unknown factors regulating synthesis of the telomeric complex itself.
Third, because telomerase activity is detected in almost all tumor and immortalized cell lines, the study of correlation of telomerase activity to oncogenesis and senescence will continue.
Since telomerase is a "hot spot" of modern molecular biology and many leading groups all over the world are involved in studies of this unusual enzyme, one can expect that in the near future our knowledge about structure, function, mechanism, and regulation of activity of telomerase will considerably increase.
LITERATURE CITED
1.McClintock, B. (1942) Proc. Natl. Acad. Sci.
USA, 28, 405-413.
2.Olovnikov, A. M. (1971) Dokl. Akad. Nauk
SSSR, 201, 1496-1499.
3.Watson, J. D. (1972) Nature New Biol.,
239, 197-201.
4.Greider, C. W., and Blackburn, E. H. (1985)
Cell, 43, 405-413.
5.Greider, C. W., and Blackburn, E. H. (1987)
Cell, 51, 887-898.
6.Greider, C. W., and Blackburn, E. H. (1989)
Nature, 337, 331-337.
7.Harley, C. B., Kim, N. W., Prowse, K. R., Weinrich,
S. L., Hirsch, K. S., West, B. M. D., Bacchetti, S., Hirte, H. W.,
Counter, C. M., and Greider, C. W. (1994) Cold Spring Harbor Symp.
Quant. Biol., 59, 307-315.
8.Counter, C. M., Hirte, H. W., Bacchetti, S., and
Harley, C. B. (1994) Proc. Natl. Acad. Sci. USA, 91,
2900-2904.
9.Kim, N. W., Piatyzek, M. A., Prowse, K. R., Harley,
C. B., West, M. D., Ho, P. L. C., Coviello, G. M., Wright, W. E.,
Weinrich, S. L., and Shay, J. W. (1994) Science, 266,
2011-2014.
10.Harley, C. B., and Villeponteau, B. (1995)
Curr. Opin. Genet. Dev., 5, 249-255.
11.Harley, C. B., Futcher, A. B., and Greider, C. W.
(1990) Nature, 345, 458-460.
12.Harley, C. B. (1991) Mutat. Res.,
256, 271-282.
13.Counter, C. M., Avilion, A. A., LeFeuvre, C. E.,
Stewart, N. G., Greider, C. W., Harkey, C. B., and Bacchetti, S. (1992)
EMBO J., 11, 1921.
14.Vaziri, H., Dragowska, W., Allsopp, R. C.,
Thomas, T. E., Harley, C. B., and Landsdorp, P. M. (1994) Proc.
Natl. Acad. Sci. USA, 91, 9857-9860.
15.Collins, K., Koybayashi, R., and Greider, C. W.
(1995) Cell, 81, 677-686.
16.Fangman, W. L., and Brewer, B. J. (1992)
Cell, 71, 363-366.
17.DeLange, T. (1994) Proc. Natl. Acad. Sci.
USA, 91, 2882-2885.
18.Zhu, X., Kumar, R., Mandal, M., Sharma, N.,
Sharma, H. W., Dhingra, U., Sokoloski, J. A., Hsiao, R., and Narayanan,
R. (1996) Proc. Natl. Acad. Sci. USA, 93, 6091-6095.
19.Cohn, M., and Blackburn, E. H. (1995)
Science, 269, 396-400.
20.Linger, J., and Cech, T. R. (1996) Proc. Natl.
Acad. Sci. USA, 93, 10712-10717.
21.Mantell, L. L., and Greider, C. W. (1994) EMBO
J., 13, 3211-3217.
22.Prowse, K. R., Avilion, A. A., and Greider, C. W.
(1993) Proc. Natl. Acad. Sci. USA, 90, 1493-1497.
23.Greider, C. W. (1995) in Telomeres
(Blackburn, E. H., and Greider, C. W., eds.) Cold Spring Harbor
Laboratory Press, New York, pp. 35-68.
24.Yu, G.-L., Bradley, J. D., Attardi, L. D., and
Blackburn, E. H. (1990) Nature, 344, 126-132.
25.Yu, G.-L., and Blackburn, E. H. (1991)
Cell, 67, 823-832.
26.Prescott, J., and Blackburn, E. H. (1997)
Genes Dev., 11, 528-540.
27.Greider, C. W. (1991) Mol. Cell Biol.,
11, 4572-4580.
28.Collins, K., and Greider, C. W. (1993) Genes
Dev., 7, 1364-1376.
29.Morin, G. B. (1989) Cell, 59,
521-529.
30.Morin, G. B. (1991) Nature, 353,
454-456.
31.Hammord, P. W., Lively, T. N., and Cech, T. R.
(1997) Mol. Cell Biol., 17, 296-308.
32.Borukhov, S., Sagitov, V., and Goldfarb, A.
(1993) Cell, 72, 459-466.
33.Jonson, T. L., and Chamberlin, M. J. (1994)
Cell, 77, 217-224.
34.Zahler, A. M., Williamson, J. R., Cech, T. R.,
and Prescott, D. M. (1991) Nature, 350, 718-720.
35.Harrington, L. A., and Greider, C. W. (1991)
Nature, 353, 451-454.
36.Lee, M. S., and Blackburn, E. H. (1993) Mol.
Cell Biol., 13, 6586-6599.
37.Wang, H., and Blackburn, E. H. (1997) EMBO
J., 16, 866-879.
38.Melek, M., Greene, E. C., and Shippen, D. E.
(1996) Mol. Cell Biol., 16, 3437-3445.
39.Collins, K., and Greider, C. W. (1995) EMBO
J., 14, 5422-5432.
40.Autexier, C., and Greider, C. W. (1994) Genes
Dev., 8, 563-575.
41.Surratt, C. K., Milan, S. C., and Chamberlin, M.
J. (1991) Proc. Natl. Acad. Sci. USA, 88, 7983-7987.
42.Izban, M. G., and Luse, D. S. (1993) J. Biol.
Chem., 268, 12864-12873.
43.Shippen-Lentz, D., and Blackburn, E. H. (1990)
Science, 247, 546-552.
44.Linger, J., Hendrick, L. L., and Cech, T. R.
(1994) Genes Dev., 8, 1984-1998.
45.Feng, J., Funk, W. D., Wang, S.-S., Weinrich, S.
L., Avilion, A. A., Chiu, C.-P., Adams, R. R., Chang, E., Allsopp, R.
C., Yu, J., Le, S., West, M. D., Harley, C. B., Andrews, W. H.,
Greider, C. W., and Villeponteau, V. (1995) Science,
269, 1236-1241.
46.Singer, M. S., and Gottschling, D. E. (1994)
Science, 266, 404-409.
47.McCormick-Graham, M., and Romero, D. P. (1995)
Nucleic Acids Res., 23, 1091-1097.
48.McEachern, M. J., and Blackburn, E. H. (1995)
Nature, 376, 403-409.
49.Blasco, M. A., Funk, W., Villeponteau, B., and
Greider, C. W. (1995) Science, 269, 1267-1270.
50.Romero, D. P., and Blackburn, E. H. (1991)
Cell, 67, 343-353.
51.Greider, C. W. (1996) Ann. Rev. Biochem.,
65, 337-365.
52.Ares, M. (1986) Cell, 47,
49-59.
53.Sogin, M. L., Ingold, A., Karlok, M., Nielsen,
H., and Engberg, J. (1986) EMBO J., 5, 3625-3630.
54.Shumyatsky, G., and Reddy, R. (1992) Nucleic
Acids Res., 20, 2159-2163.
55.Bhattacharyya, A., and Blackburn, E. H. (1994)
EMBO J., 13, 5721-5731.
56.Gilley, D., Lee, M. S., and Blackburn, E. H.
(1995) Genes Dev., 9, 2214-2226.
57.Gilley, D., and Blackburn, E. H. (1996) Mol.
Cell Biol., 16, 66-75.
58.Bhattacharya, A., and Blackburn, E. H. (1997)
Proc. Natl. Acad. Sci. USA, 94, 2823-2827.
59.Green, R., and Noller, H. F. (1996) RNA,
2, 1011-1021.
60.Noller, H. F. (1984) Ann. Rev. Biochem.,
53, 119-162.
61.Nierhaus, K. H. (1991) Biochimie,
73, 739-755.
62.Autexier, C., Pruzan, R., Funk, W. D., and
Greider, C. W. (1996) EMBO J., 15, 5928-5935.
63.Linger, J., Hughes, T. R., Shevchenko, A., Mann,
M., Lundblad, V., and Cech, T. R. (1997) Science, 276,
561-567.
64.Lundblad, V., and Szostak, J. W. (1989)
Cell, 57, 633-643.
65.Lundblad, V., and Blackburn, E. H. (1990)
Cell, 60, 529-530.
66.Lundblad, V., and Blackburn, E. H. (1993)
Cell, 73, 347-360.
67.Lin, J.-J., and Zakian, V. A. (1995) Cell,
81, 1127-1135.
68.Steiner, B. R., Hidaka, K., and Futcher, B.
(1996) Proc. Natl. Acad. Sci. USA, 93, 2817-2821.
69.Virta-Pearlman, V., Morris, D. K., and Lundblad,
V. (1996) Genes Dev., 10, 3094-3104.
70.Lendvay, T. S., Morris, D. K., Sah, J.,
Balasubramanian, B., and Lundblad, V. (1996) Genetics,
144, 1399-1412.
71.Nakayama, J., Saito, M., Nakamura, H., Matsuura,
A., and Ishikawa, F. (1997) Cell, 88, 875-884.
72.Harrington, L., McPhail, T., Mar, V., Zhou, W.,
Oulton, R., Bass, M. B., Arruda, I., and Robinson, M. O. (1997)
Science, 275, 973-977.
73.Neer, E. J., Schmidt, C. J., Nambudrioad, R., and
Smith, T. F. (1994) Nature, 371, 297-300.
74.Hiyama, K., Yokoyama, T., Matsuura, Y.,
Piatyszek, M. A., and Shay, J. W. (1995) Nature Med.,
1, 249-255.
75.Broccoli, D., Young, J. W., and de Lange, T.
(1995) Proc. Natl. Acad. Sci. USA, 92, 9082-9086.
76.Bryan, T. M., Engelzou, A., Gupta, J., Bacchetti,
S., and Reddel, R. R. (1995) EMBO J., 14, 4240-4244.
77.Klingelhutz, A. J., Foster, S. A., and McDougall,
J. K. (1996) Nature, 380, 79-81.
78.Norton, J. C., Piatyszek, M. A., Wright, W. E.,
Shay, J. W., and Corey, D. R. (1996) Nature Biotech.,
14, 615-619.
79.Strahl, C., and Blackburn, E. H. (1996) Moll.
Cell Biol., 1, 53-65.
80.Yegorov, Y. E., Chernov, D. N., Akimov, S. S.,
Bolsheva, N. L., Kraevsky, A. A., and Zelenin, A. V. (1996) FEBS
Lett., 389, 115-118.